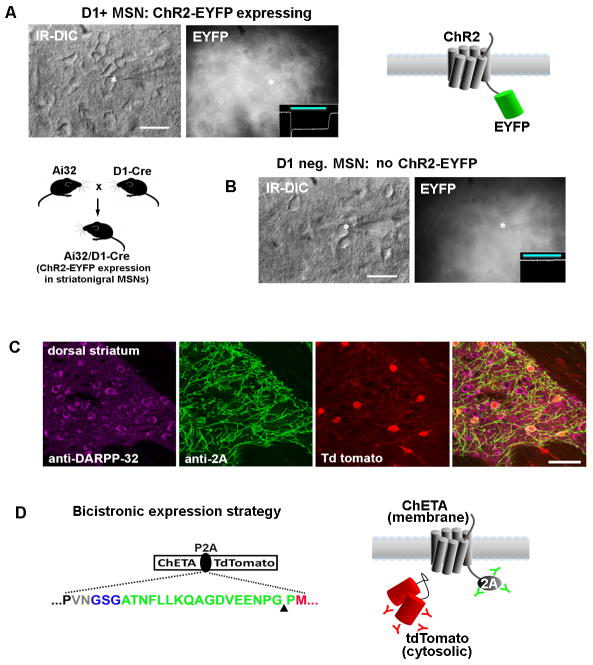
Figure 6.
Improved visualization of ChR2-expressing neurons for targeted patch clamp recordings in brain slices using viral P2A linkers.(A) The challenge of identifying ChR2-EYFP expressing neurons is examined in acute striatal brain slices from Ai32/D1-Cre mice. This line has strong expression of the ChR2-EYFP transgene in roughly half of all striatal medium spiny neurons. The ChR2-EYFP gene fusion is localized to the cell membrane and produces a dense fluorescent neuropil with little signal from cell bodies. A recorded neuron is shown (asterisk) along with an inset of recorded photocurrent, thus confirming the identity as a D1+ MSN.(B) A recorded MSN in a nearby region had no photocurrent and was presumed D1 negative (D1−/D2+ MSNs account for the other half of the MSN population). The recorded neurons were indistinguishable on the basis of morphology or live EYFP fluorescence.(C) Analysis of native tdTomato fluorescence together with double immunostaining with anti-2A (indicating localization of ChR2) and DARPP32 (indicating all MSNs in the striatum region) demonstrates the unambiguous identification of ChR2 expressing neurons with the opsin-2A-XFP expression strategy. The example shown here is from our Cre-inducible ChETA-P2A-tdTomato reporter line crossed to the RGS9-Cre driver mice for labeling a subset of striatal MSNs.(D) The use of the viral P2A linker (green sequence) allows for physical uncoupling of opsin and fluorophore, and the 2A epitope tag can then be used to track the localization of the membrane-targeted opsin protein. The cytosolic fluorophore, in this case tdTomato, fills the entire cell body. Scale bars: 20 μm in panels A and B, 50 μm in panel C.