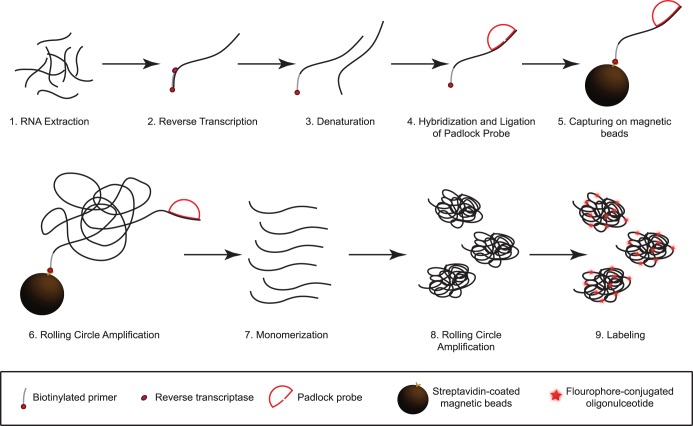
Figure 2. Schematic summary of assay principle.
(1) RNA is extracted from patient samples and (2) cDNA is synthesized using biotinylated primers. (3) The DNA is denatured and (4) padlock probes are hybridized and ligated to their complementary target sequence. (5) This complex is captured onto magnetic beads to wash away unbound probes and (6) amplified by rolling circle amplification (RCA) for 20 min. (7) The rolling circle products (RCPs) are monomerized and (8) a second round of RCA is performed. (9) Fluorescently labeled oligonucleotides are hybridized to the RCPs for detection in a microfluidic setup using a confocal microscope.