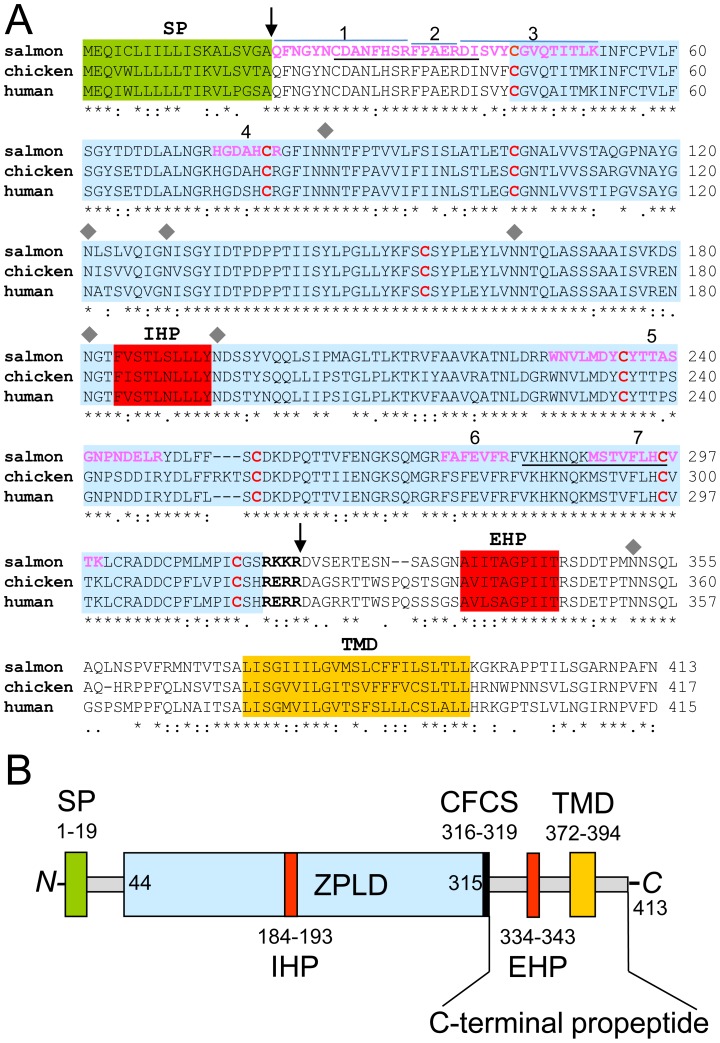
Figure 3. Zona pellucida-like domain protein homology and structure.
A, protein sequences from UniProt database: salmon: C0H9B6; chicken: E1C8E6; human: Q8TCW7 were aligned by applying the ClustalW2 program. Asteriks (★) marked below the sequence highlight conserved amino-acids (∼70%) between the three organisms. Conserved cysteine residues 1–8 that constitute the zona pellucida-like domain (blue box) are shown in red letters. Arrows mark the mature protein after cleavage of the N-terminal signal sequence (SP, green box) predicted by the SignalP algorithm and the C-terminal furin cleavage site (CFCS, black and bold letters). IHP and EHP (red boxes) show the potential internal and external hydrophobic patches of the zona pellucida-like-domain (ZLPD). The transmembrane domain (TMD, orange box) was predicted by the TMHMM 2.0 software. In pink and bold are highlighted the peptides 1–7, identified by mass spectrometry. Individual peptides 1, 2 and 3 are highlighted with blue lines. Underlined peptide sequences were used for immunization. Grey diamonds indicate the asparagine residue of potential N-glycosylation sites (NXS/T) determined with the program NetNGlyc 1.0. B, scheme of zona pellucida-like domain protein structure.