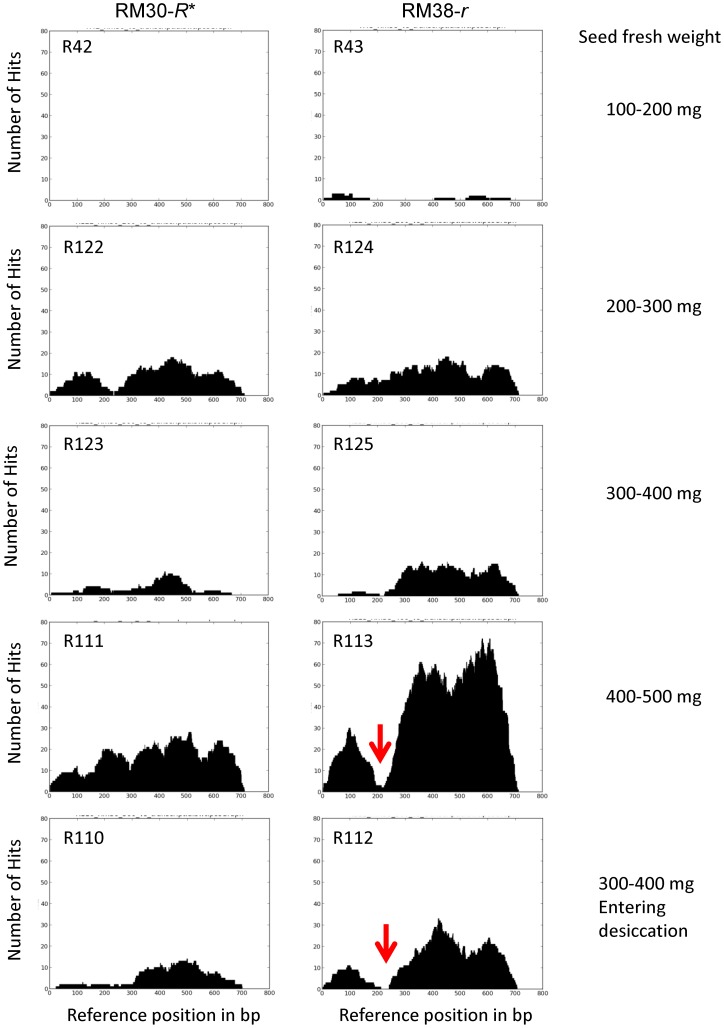
Figure 9. Distribution of RNA-Seq Reads from Seed Coats of the RM30-R* and RM38-r When Aligned to the Transcript Sequence of Glyma09g36983.
The pair of graphs in each row represent the distribution of non-normalized RNAseq reads that align to the 714 bp transcript sequence from the Glyma09g36983 gene at each of the five stages of seed coat development indicated in the margin to the right (described in Figure 4). The plots in the left column represent the distributions of sequence reads derived from the seed coats of the RM30-R* that is interrupted by the TgmR* transposon and the plots in the right column represent the RNAseq reads derived from the RM38-r line with the uninterrupted R gene but containing the “C”-nt deletion at 222 bp of the transcript. The numbers of RNAseq reads matching a given position of the R gene transcript are plotted against the position along the reference Glyma09g36983 transcript sequence containing the three exons and no introns. No expression was detected in the early 100–200 mg seed developmental stage in the RM30-R* line. The red arrow points to the location of the r allele “C”-nt deletion at 222 bp of the transcript.