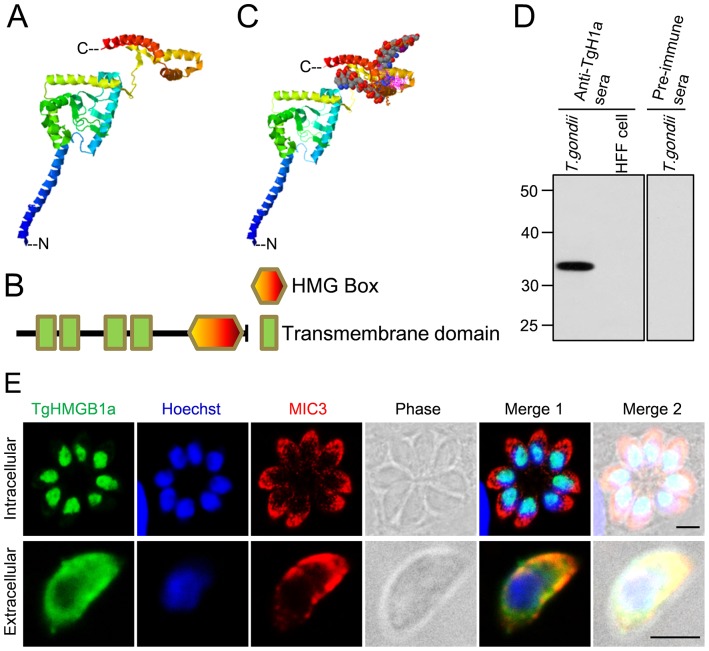
Figure 2. Characterization of TgHMGB1a by structure modeling and cellular localization.
A. Ribbon cartoon representations of the predicted 3D structure of TgHMGB1a generated by homology modeling. Three α-helices independently fold into the HMG box at the C-terminal, while another four helices are clustered as the transmembrane domains. C indicated the C-terminal and N showed the N-terminal. B. Schematic representation of the primary structure of TgHMGB1a highlighting its different domains. TgHMGB1a has only one HMG box at the C-terminus and four transmembrane domains at the N- terminal; there is no acidic C-tail. C. A model of TgHMGB1a bound to DNA. TgHMGB1a can bind DNA through an interaction between the HMG box and the minor groove of DNA. D. Identification of native TgHMGB1a by western blotting. T. gondii represents the total antigens from cell cultured T. gondii tachyzoites; uninfected HFF cell lysates were used as a control. A specific band was elicited when anti-rTgHMGB1a 4E polyclonal antibodies were used as a primary antibody, but not when preimmune serum was used. E. TgHMGB1a is concentrated in the nucleus of parasite. Immunofluorescent localization of TgHMGB1a showed that TgHMGB1a (green) not only localized in the nucleus (blue) during cytokinesis (C) (top panel), but also during mitosis (M) (middle panels). Additionally, TgHMGB1a continued to accumulate in the nucleus of tachyzoites freshly released into extracellular media (bottom panel). MIC3 (red), nuclei stained with Hochest (blue). Scale bars, 3 µm. All immunofluorescent labeling was performed on HFF cells infected with tachyzoites of the RH strain.