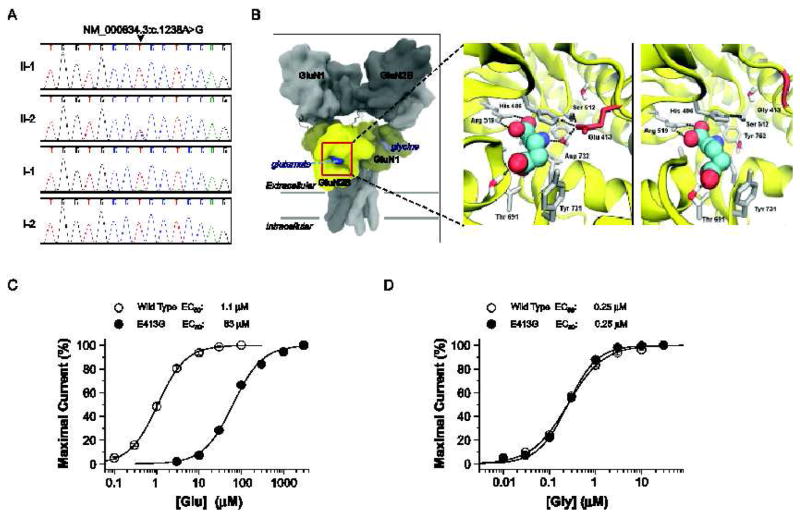
Fig. 4.
Identification and characterization of a de novo GRIN2B (GluN2B) mutation in patient 2. (A) Sanger sequence traces showing a heterozygous GRIN2B mutation (NM_000834.3:c.1238A>G, NP_000825.2:p.E413G) only in patient 2 (II-2). (B) Predicted quaternary structure of tetrameric human GluN1/GluN2B, based on the GluA2 AMPA receptor structure (left; 3KG2) [52]. The bi-lobed ligand binding domain (yellow) of GluN2B and related glutamate receptors adopts a clamshell-like structure, which binds glutamate (blue) within the cleft. Ligand-protein interactions between L-glutamate (spheres) and wild-type GluN2B (middle) or GluN2B-E413G (right) ligand binding domains, are modeled from GluN1/GluN2A crystallographic data (2A5T) [15]. Nearby residues and crystallographically conserved waters are shown (sticks). Hydrogen bonds are depicted by the black dashed lines. (C) The composite concentration-response curves and fitted EC50 values are shown for human GluN1/GluN2B (wild type) or GluN1/GluN2B-E413G (E413G) current responses (100 μM glycine in all solutions, VHOLD −40 mV). (D) The composite glycine concentration-response curves and fitted EC50 values at human GluN1/GluN2B (wild type) or GluN1/GluN2B-E413G (E413G) receptors (1 mM glutamate present in all solutions). EC50 values were obtained by fitting the curves in (C) and (D) with Response (%) = 100 / (1 + (EC50 / [agonist])N). N is the Hill slope, which ranged between 1.1 – 1.4; n = 11–20 oocytes).