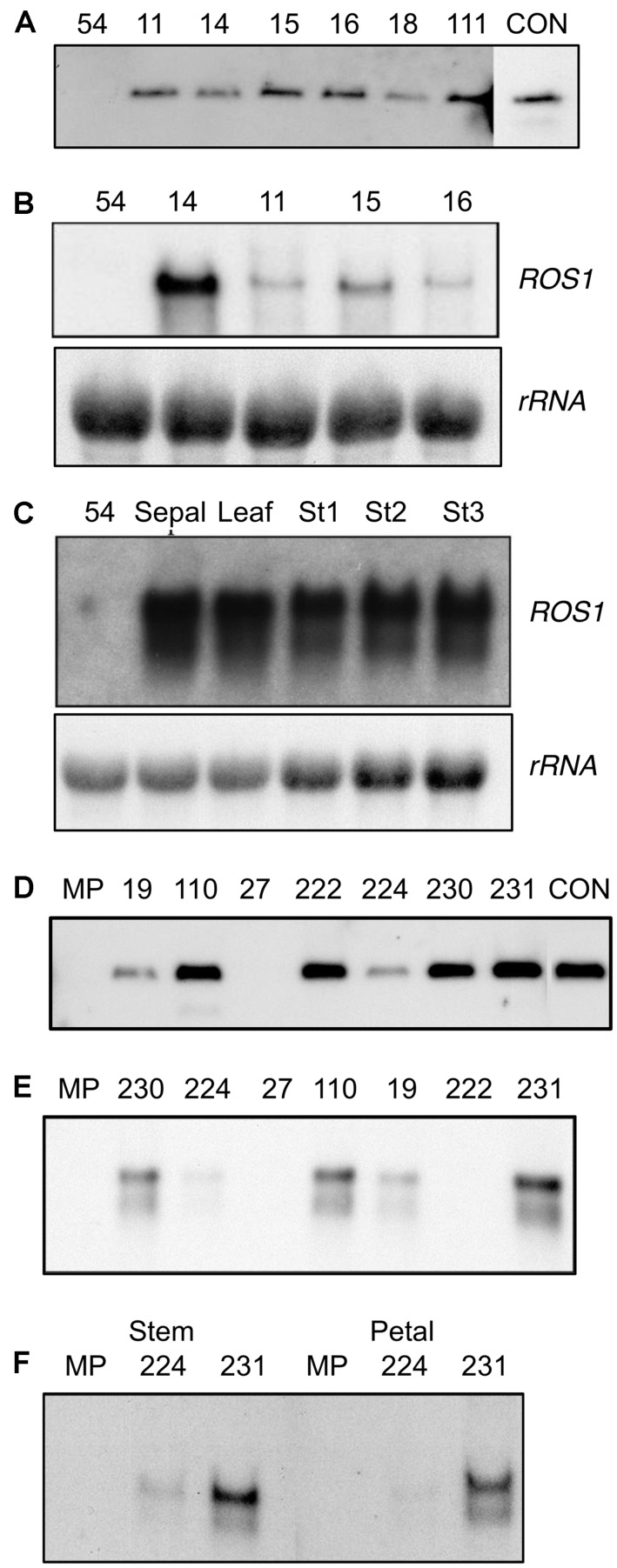
FIGURE 2.
Analysis of 35S:ROS1 plants for the presence and expression of the transgene. (A) Southern hybridization of 35S:ROS1 lisianthus plants using radiolabelled ROS1 cDNA as the probe. Thirty μg of genomic DNA was digested with the restriction enzyme Asp718 to release the ROS1 fragment from the T-DNA. The control (CON) lane contained pLN81-derived DNA and shows the expected fragment from an Asp718 digest. The lane labeled ‘54’ contains genomic DNA from a non-transgenic lisianthus cv. 54 plant. (B) Northern hybridization analysis of ROS1 transcript abundance in petal RNA of a non-transgenic control plant (54) and four lines of 35S:ROS1 lisianthus (14, 11, 15, 16). (C) Northern hybridization analysis of ROS1 transcript abundance in sepal, leaf, and three petal stages (Stage 1,3 and 5) of 35S:ROS1 lisianthus line 54-14 and petal RNA of a non-transgenic control plant (54). In both (B) and (C) 20 μg of total RNA was hybridized with radiolabelled ROS1 cDNA or an rRNA cDNA (for an estimation of RNA loading) as the probe. (D) Southern hybridization of 35S:ROS1 MP petunia plants using radiolabelled ROS1 cDNA as the probe. Thirty μg of genomic DNA was digested with the restriction enzyme Asp718 to release the ROS1 fragment from the T-DNA. The control (CON) lane contained pLN81-derived DNA and shows the expected fragment from an Asp718 digest. The lane labeled ‘MP’ contains genomic DNA from a non-transgenic MP plant. (E) Northern hybridization analysis of ROS1 transcript abundance in total RNA (10 μg) from petals of seven 35S:ROS1 MP plants and a non-transgenic control (MP). RNA was hybridized with radiolabelled ROS1 cDNA as the probe. (F) Northern hybridization analysis of ROS1 transcript abundance in total RNA from stem (25 μg) or petal (30 μg) RNA from 35S:ROS1 lines MP224 and MP231 and a non-transgenic control (MP).