Fig 6.
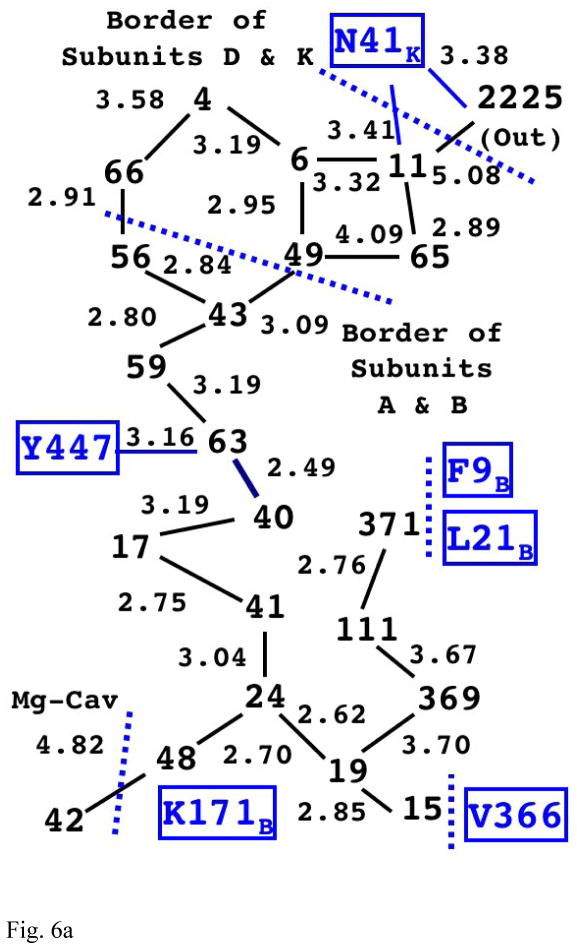
Fig. 6a The Structure of Internal Water Molecules in Channel-R of Mitochondrial CcO (1V55)
The integers represent the water molecules observed in the x-ray structure (with the value indicating their residue index number in the original PDB file). The lines between them show the possible hydrogen bond links between them with the distance (in Å) shown on the side. Note that there is a complete chain of hydrogen bonding (with gaps less than 3.4Å) from water 48 all the way up to water 11. (Also see Fig. S1 in Supplementary Information for actual positioning of the water molecules with the cavity.)
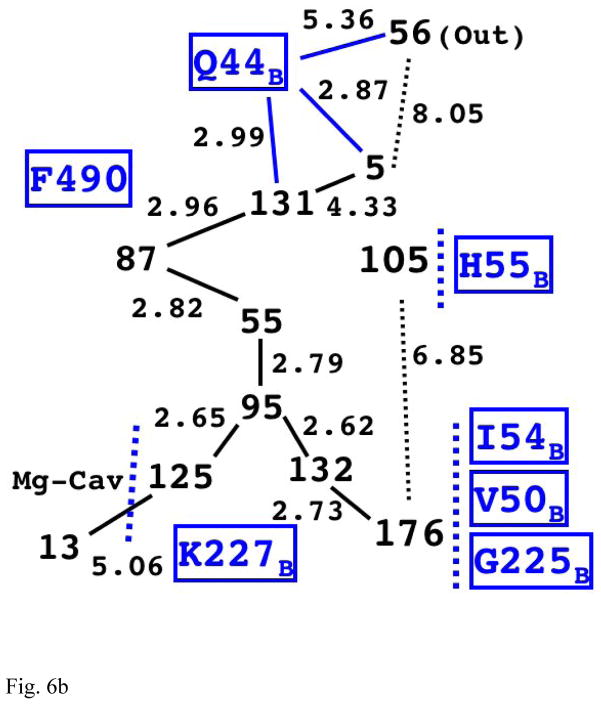
Fig. 6b The Structure of Internal Water Molecules in Channel-R for CcO of Rhodobacter sphaeroides (2GSM)
The first four water molecules on the longest chain (125, 95, 55, and 87) are structurally very similar to the corresponding chain in the eukaryotic enzyme (Fig. 6a).
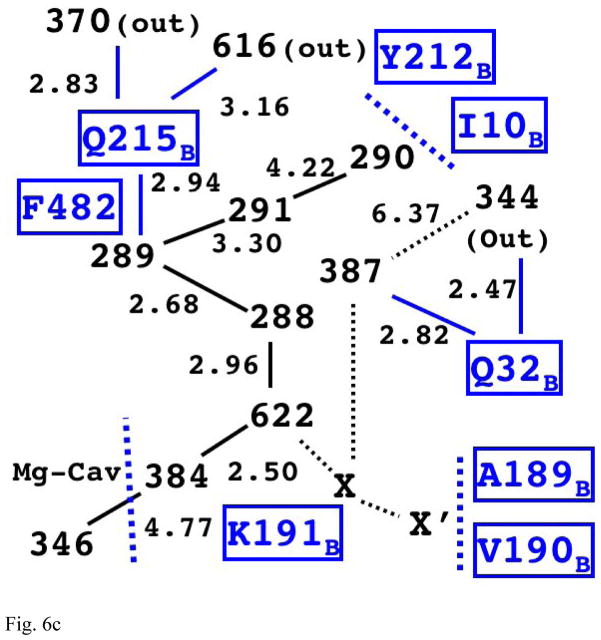
Fig. 6c The Structure of Internal Water Molecules in Channel-R for CcO of Paracoccus denitrificans (3EHB, N131D mutant)
The relative positions of water molecules on the main chain is equivalent to the one in Rhodobacter sphaeroides; however, it appears to have a different possible exit for proton via Q215B.