Table 1. Statistics of data collection and refinement.
Values in parentheses are for the highest resolution shell.
| Data set | Csd4-unbound | Csd4muramyltripeptide | Csd4mDAP | SeMet (peak) |
|---|---|---|---|---|
| Data collection | ||||
| Beamline source† | PF BL-17A | PLS BL-7A | PLS BL-7A | PLS BL-7A |
| X-ray wavelength () | 1.0722 | 0.9800 | 1.2823 | 0.9795 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| a, b, c () | 53.14, 66.55, 144.05 | 53.13, 66.38, 143.79 | 52.93, 66.67, 144.00 | 53.17, 66.57, 144.39 |
| Resolution range () | 30.01.60 (1.631.60) | 50.01.55 (1.581.55) | 50.01.41 (1.431.41) | 30.01.60 (1.631.60) |
| Total No. of reflections | 686326 (33531) | 375793 (19100) | 475587 (19787) | 1655884 (81493)‡ |
| No. of unique reflections | 68532 (3387) | 71540 (3537) | 94978 (4497) | 131002 (6572)‡ |
| Multiplicity | 10.0 (9.9) | 5.3 (5.4) | 5.0 (4.4) | 12.6 (12.4)‡ |
| Completeness (%) | 99.9 (100.0) | 95.7 (96.1) | 95.7 (91.9) | 100.0 (100.0)‡ |
| I/(I) | 47.0 (5.7) | 31.5 (4.1) | 36.6 (2.1) | 39.5 (4.6)‡ |
| Wilson B factor (2) | 27.0 | 24.3 | 25.1 | 24.9 |
| R merge § (%) | 7.0 (53.1) | 8.5 (58.9) | 7.7 (75.8) | 10.1 (68.7)‡ |
| R r.i.m. ¶ (%) | 7.4 (56.0) | 9.5 (65.2) | 8.5 (85.5) | 10.5 (71.6)‡ |
| R p.i.m. †† (%) | 2.3 (17.7) | 4.0 (27.3) | 3.7 (38.6) | 3.0 (20.2)‡ |
| SAD phasing | ||||
| Figure of merit (before/after density modification) | 0.39/0.64 | |||
| Model refinement | ||||
| PDB code | 4q6m | 4q6n | 4q6o | |
| No. of Csd4 monomers in asymmetric unit | 1 | 1 | 1 | |
| Resolution range () | 20.01.60 | 20.01.55 | 20.01.41 | |
| R work/R free ‡‡ (%) | 19.4/22.0 | 20.0/22.7 | 18.9/21.2 | |
| No. of non-H atoms | ||||
| Total | 3812 | 3860 | 4114 | |
| Protein | 3366 | 3366 | 3366 | |
| Water O | 437 | 446 | 708 | |
| Glycerol | 6 | 18 | 24 | |
| Calcium ion | 3 | 3 | 3 | |
| Muramyltripeptide | 27 | |||
| mDAP | 13 | |||
| Average B factor (2) | ||||
| Overall | 20.2 | 19.6 | 23.2 | |
| Protein | 18.9 | 18.1 | 19.9 | |
| Water O | 30.1 | 29.8 | 38.0 | |
| Glycerol | 39.5 | 39.9 | 48.5 | |
| Calcium ion | 16.3 | 17.5 | 2.3 | |
| Muramyltripeptide | 30.1 | |||
| mDAP | 19.9 | |||
| R.m.s. deviations from ideal geometry | ||||
| Bond lengths () | 0.009 | 0.008 | 0.007 | |
| Bond angles () | 1.35 | 1.32 | 1.31 | |
| R.m.s. Z-scores§§ | ||||
| Bond lengths | 0.435 | 0.404 | 0.339 | |
| Bond angles | 0.618 | 0.596 | 0.596 | |
| Ramachandran plot¶¶ (%) | ||||
| Favoured/outliers | 97.4/0.0 | 97.6/0.0 | 97.7/0.0 | |
| Poor rotamers¶¶ (%) | 0.3 | 0.8 | 1.0 | |
PF, Photon Factory, Japan; PLS, Pohang Light Source, Republic of Korea.
Friedel pairs were treated as separate observations.
R
merge = 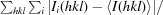
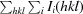 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 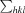 is the sum over all reflections and
is the sum over all reflections and 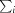 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
R
r.i.m. = 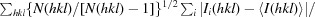
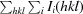 .
.
R
p.i.m. = 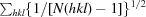
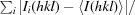
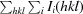 .
.
R
work = 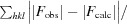
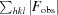 , where R
free is calculated for a randomly chosen 5% of reflections which were not used for structure refinement and R
work is calculated for the remaining reflections.
, where R
free is calculated for a randomly chosen 5% of reflections which were not used for structure refinement and R
work is calculated for the remaining reflections.
Values were obtained using REFMAC.
Values were obtained using MolProbity.
