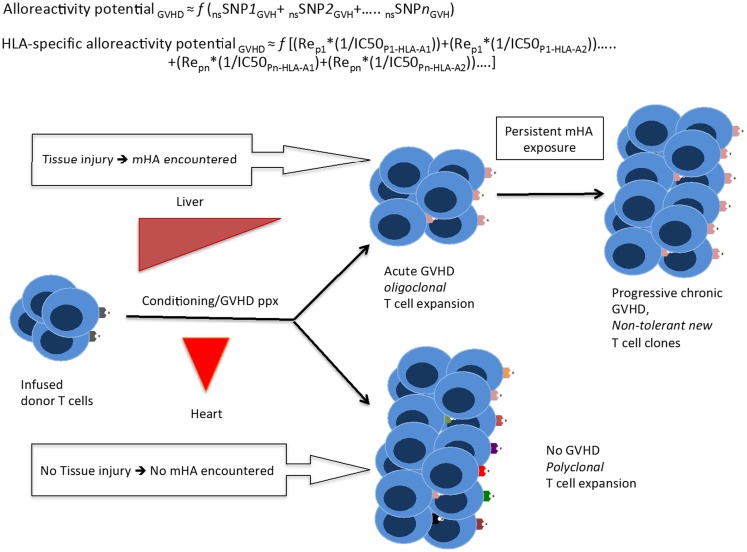
Figure 7.
A quantitative model for the development of GVHD. Whole exome sequencing identifies all the nsSNP with a GVH vector, yielding a putative alloreactivity potential, which may be a function (f) of the cumulative influence of these polymorphisms. This is represented as a series, listing the sequence of polymorphic exome loci. Substituting individual nsSNPGVH in the equation by peptide-HLA binding affinity (reciprocal of IC50)*relative expression level of the gene bearing the nsSNPGVH (for each HLA molecule) yields the HLA-specific alloreactivity potential, in this Re is the relative expression of protein with nsSNPGVH and resulting peptides. In this series, the expression, Rep1*(1/IC50P1-HLA-A1) for each specific peptide-HLA complex, hypothetically represents the T-cell clone-specific AP. Multiple peptides constituting this series then drive a proportional oligoclonal T-cell expansion in GVHD, as many different mHA are presented by the HLA in an individual, the final distribution conforming to the Power law. Since T-cell clonal expansion in response to presented antigens may be influenced by factors such as tissue injury, cytokine milieu, and immunosuppression intensity; the GVHD likelihood, and its phenotype may in turn be determined not only by the ubiquitous mHA but also by the tissue volume and its state (inflammation/injury), and most importantly time at which organ injury/inflammation occurs relative to T-cell infusion.