Figure 1.
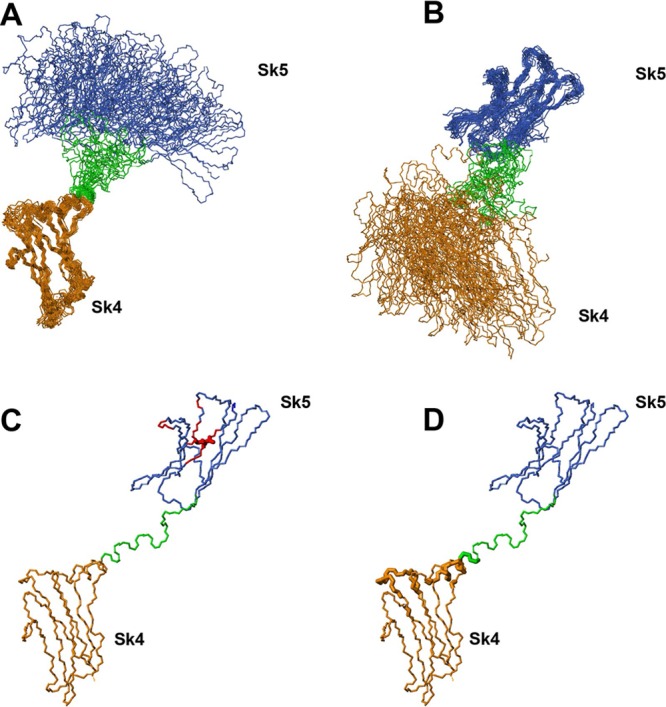
Sk45 NMR ensemble obtained through the calculation where RDC data for the two domains were used independently (A, B). Sk4 domain and N-terminal part of the linker is shown in orange, Sk5 domain–in blue, the unstructured part of the linker–in green. (A) The structures are superimposed over the domain 4 of the tandem Sk45 well-ordered residues 1227–1327. (B) The structures are superimposed over domain 5 of the tandem Sk45 well-ordered residues 1344–1380 and 1391–1427. Additional data used to confirm interdomains orientation (C–D). (C) The residues affected by the introduction of m-PROXYL spin label are in red. The spin modified residue is shown with the thicker bonds and side chain displayed. (D) The residues of Sk4 domain and the beginning of linker affected by the presence of the Sk5 domain are shown with thicker bonds. The affected residues were deduced from chemical shift perturbations in 1H–15N HSQC spectra of Sk4 and Sk45 constructs.
