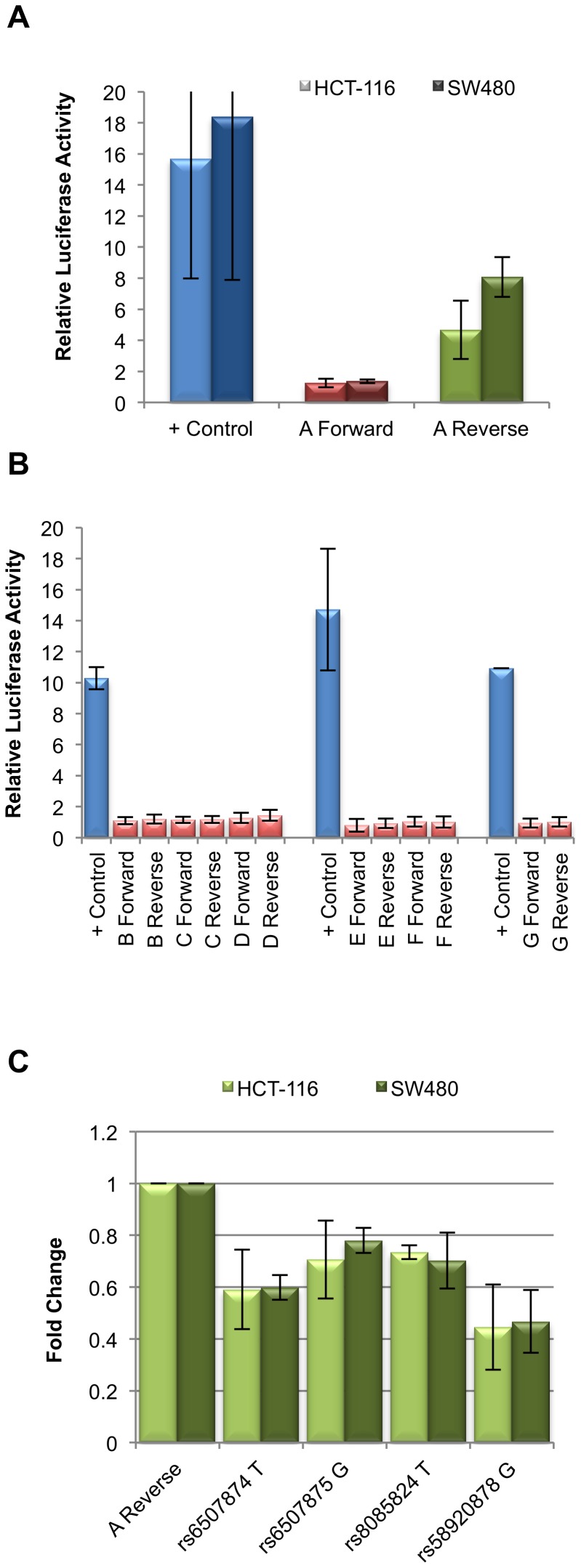
Figure 2. Fragment A exhibits enhancer activity in the reverse orientation only.
(A) Relative luciferase activity for positive control, fragment A in the forward, and fragment A in the reverse orientation. Light columns indicate activity in HCT-116, dark columns indicate activity in SW480. (B) Fragments B through G, shown in figure 1B as red stripes do not show enhancer activity in either orientation in HCT-116. Positive controls are shown for each group of constructs analyzed on the same plate. Fragment E contains the tagSNP rs4939827. (C) Activity of fragment A in the reverse orientation with all SNPs in the fragment with their C allele, with each allele changed independently to the alternate allele, shown as fold change relative to the construct containing the four C alleles. All four alleles result in a statistically significant reduction in activity: rs6507874 T p = 8.25×10−3 (HTC-116), p = 3.30×10−2 (SW480); rs6507875 G p = 3.40×10−2 (HCT-116), p = 6.79×10−3 (SW480); rs8085824 T p = 1.20×10−2 (HCT-116), p = 3.12×10−3 (SW480); rs58920878 G p = 2.09×10−3 (HCT-116), p = 3.05×10−3 (SW480).