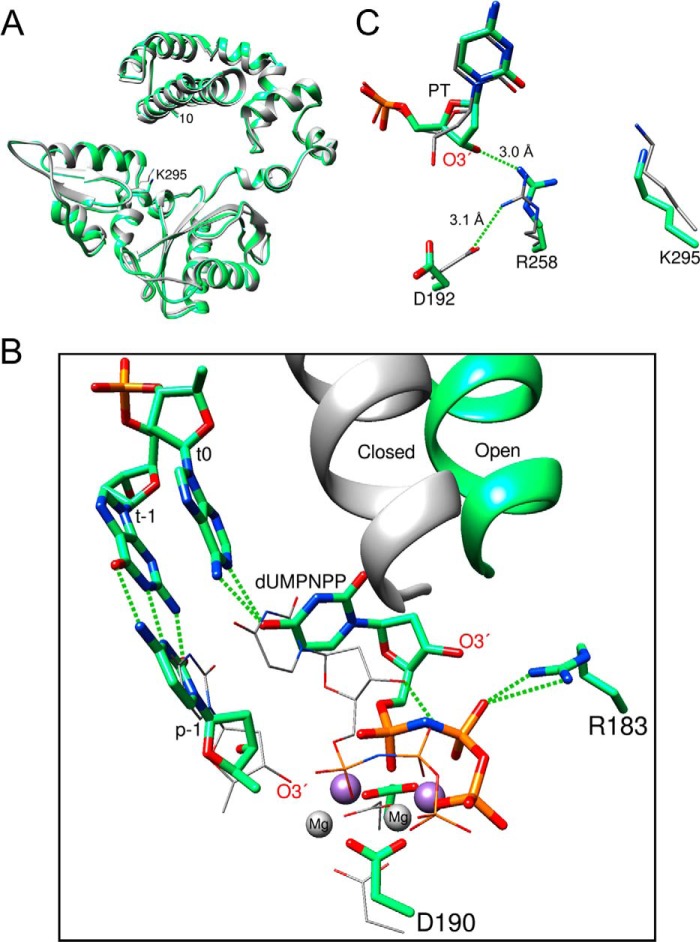
FIGURE 7.
Crystallographic structures of binary and ternary substrate complexes of the E295K mutant. A, ribbon representation of the superimposed structures of the binary DNA (gray) and ternary (green; +dUMPNPP) structures. The superimposed structures (r.m.s.d. = 0.64 Å; 322 Cα) indicate that the ternary substrate complex is in the open conformation typically observed for binary DNA complexes of pol β. The position of the Lys-295 (K295) side chain is indicated, and the first residue (residue 10) that can be observed in the structure is labeled (10). The DNA and dUMPNPP are omitted for clarity. B, active site structural comparison of the ternary substrate complexes of E295K mutant (green stick representation) and wild-type (thin gray stick representation) enzyme (PDB ID 2FMS). The open/closed position of the α-helix N is shown for these enzymes (r.m.s.d. = 0.84 Å, 201 Cα). The templating nucleotide (t0) and its upstream neighbor (t-1) are shown for the mutant enzyme, whereas the primer terminus (p-1) is shown for both the mutant and wild-type enzymes. The magnesium ions in the wild-type enzyme are shown as gray spheres, and the manganese ions of the mutant complex are shown as purple spheres. The triphosphate moiety of the incoming dUMPNPP in the mutant structure hydrogen bonds to Arg-183 (R183), whereas the O4 (uracil) is within hydrogen bonding distance to N1 and N6 of the templating adenine. C, key active site residues of the superimposed binary (thin gray stick representation) and ternary (green stick representation) complexes of the E295K mutant are shown. The primer terminus (PT) of the ternary complex is geometrically distorted removing O3′ from a catalytically relevant position. This inactive sugar position is stabilized with a hydrogen bond with Arg-258.