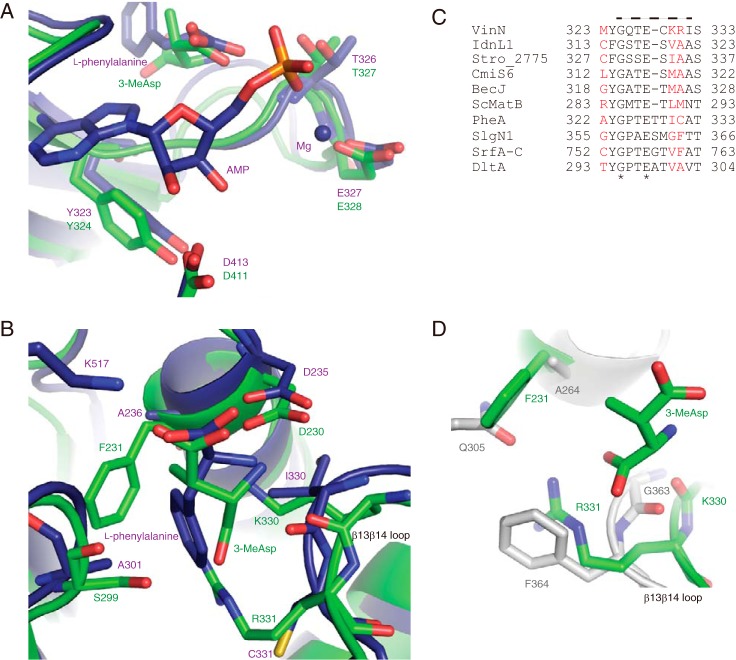
FIGURE 4.
Structural comparison of VinNN with other adenylation enzymes. A, the structure of the adenylate-binding site. The superimposed structures of VinNN (green) and PheA (purple) are shown. B, the structure of the substrate-binding pocket. The superimposed structures of VinNN (green) and PheA (purple) are shown. C, the alignment of the β13β14 loop region of VinN with other adenylation enzymes. The sequences of the following 10 adenylation enzymes were used for the alignment analysis: α-amino acid-activating enzymes, including Bacillus brevis PheA (2), Streptomyces lydicus SlgN1 (26), Bacillus subtilis SrfA-C (45), and Bacillus cereus DltA (35); β-amino acid-activating enzymes, including VinN, Streptomyces sp. ML694-90F3 IdnL1 (18), Salinispora tropica Stro_2775 (21), Streptomyces sp. MJ635-86F5 CmiS6 (19), and Streptomyces sp. DSM 21069 BecJ (20); and malonyl-CoA synthetase Streptomyces coelicolor ScMatB (44). The specificity-conferring code residues are shown in red. The VinN β13β14 loop is indicated by a broken line above the sequence. The conserved positions are marked with an asterisk. D, the superimposed substrate-binding pocket structures of VinNN (green) and SlgN1 (gray).