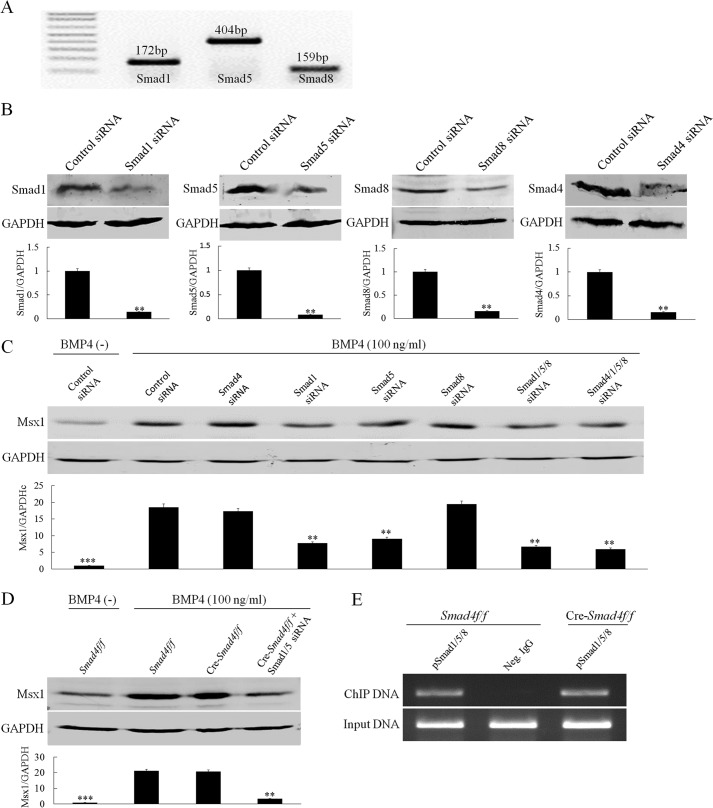
FIGURE 6.
Smad1/5 but not Smad4 are required for BMP-induced Msx1 expression in dental mesenchymal cells. A, RT-PCR shows the mRNA expressions of Smad1, Smad5, and Smad8 in E13.5 molar germs. B, Western blot shows knockdown efficiency of each siRNAs on their respective target genes in primary dental mesenchymal cells from E13.5 embryos. Quantitative analyses are shown in the lower part of each panel. C, a representative Western blot assay shows Msx1 expression in primary dental mesenchymal cells from E13.5 embryos. These cells were transfected with Smad1, Smad5, Smad8, or Smad4 siRNAs or control siRNA for 48 h followed by BMP4 treatment for additional 6 h. GAPDH was used as internal control. Quantitative analysis is shown in the lower part, in which the average value of Msx1/GAPDH from each individual group was compared with that from BMP4-induced control siRNA group. D, Western blot shows Msx1 protein levels in E13.5 Smad4f/f or Cre-Smad4f/f dental mesenchymal cells with or without transfection of Smad1/5 siRNAs after BMP4 induction for 6 h. Quantitative analysis is shown in the lower part, in which the average value of Msx1/GAPDH from Smad4f/f cells after BMP stimulation is set as the reference. E, a ChIP assay shows binding of pSmad1/5/8 to the Msx1 promoter in E13.5 dental mesenchymal cells in either the presence or the absence of Smad4. Neg. IgG, negative control IgG. **, p < 0.01; ***, p < 0.001. Error bars indicate mean ± S.D.