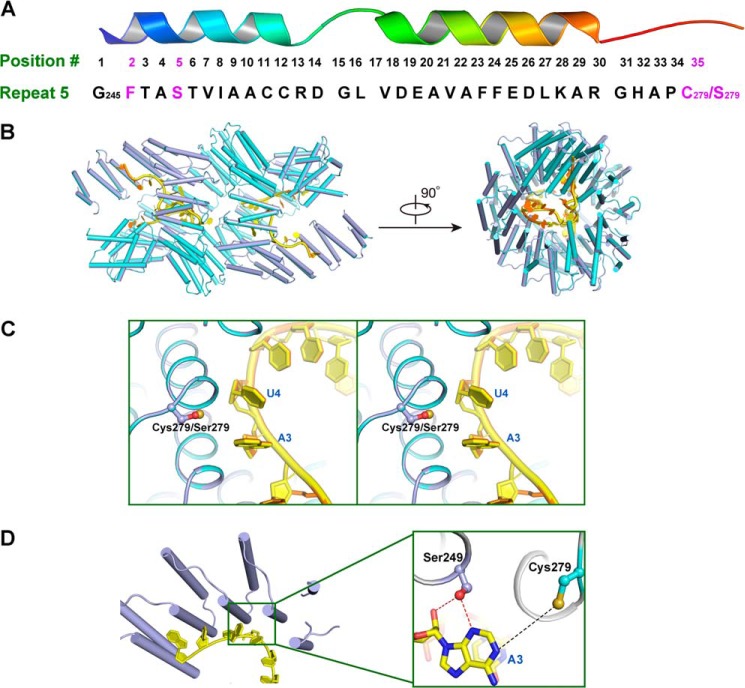
FIGURE 6.
Dimeric crystal structure of PPR10 (C256S/C430S/C449S) with RNA. A, amino acid residues in repeat 5 of PPR10 spatially organized into a hairpin of α-helices. Residues Phe-246, Ser-249, and Cys-279/Ser-279 involved in recognition of adenine base in psaJ RNA in the crystal structure are highlighted in magenta. B, superimposition of PPR10 (C256S/C430S/C449S) colored light purple with RNA in yellow and PPR10 (C256S/C279S/C430S/C449S) colored cyan with RNA in orange (Protein Data Bank code 4M59) revealed almost identical recognition pattern toward RNA. The left and right panels are related by a 90° rotation around a horizontal axis. C, regional structure comparison between PPR10 (C256S/C279S/C430S/C449S) and PPR10 (C256S/C430S/C449S) in the vicinity of Cys-279/Ser-279 residue. The side chain of Cys-279/Ser-279 is colored by atoms with sulfur in Cys-279 colored beige and oxygen in Ser-279 colored red. Stereo view of superimposition is shown. D, recognition interface between PPR10 (C256S/C430S/C449S) and RNA. A close-up view of residues in repeat 5 recognizing adenine in psaJ RNA is depicted on the right. Polar contacts and van der Waals interactions are shown as red and black dotted lines, respectively.