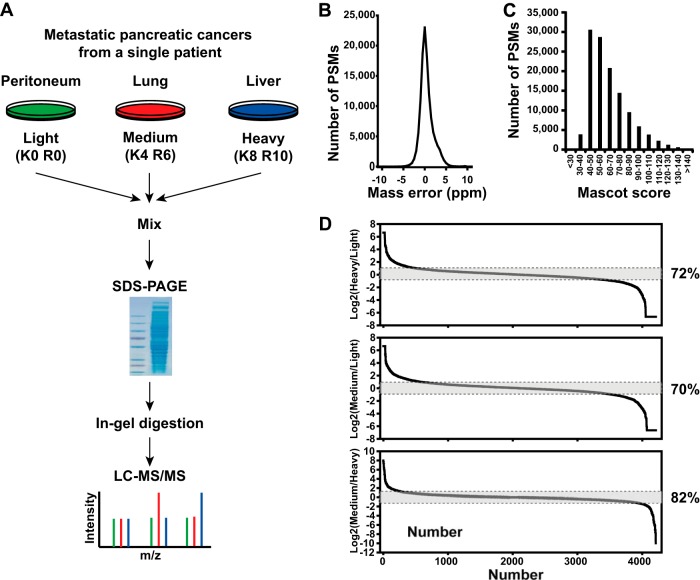
Fig. 1.
A SILAC strategy combined with high-resolution mass spectrometry for differential proteome analysis. A, schematic of a SILAC-based quantitative proteomics for differential proteome analysis of cells isolated from metastatic pancreatic cancer in distal organs. B, highly accurate mass measurements were carried out with sub-ppm error on the LTQ-Orbitrap Elite mass spectrometer. C, distribution of Mascot scores of all peptide-spectrum matches at 1% false discovery rates shows a median Mascot score of ∼59 derived from high-quality tandem mass spectrometry data acquired in this study. D, over 4,200 proteins were observed in this experiment. When two metastatic pancreatic cancer proteomes (i.e. peritoneal metastasis versus liver metastasis) were compared, ∼28% of proteins were found to vary by >2-fold among metastases, although most of the proteins remained unchanged.