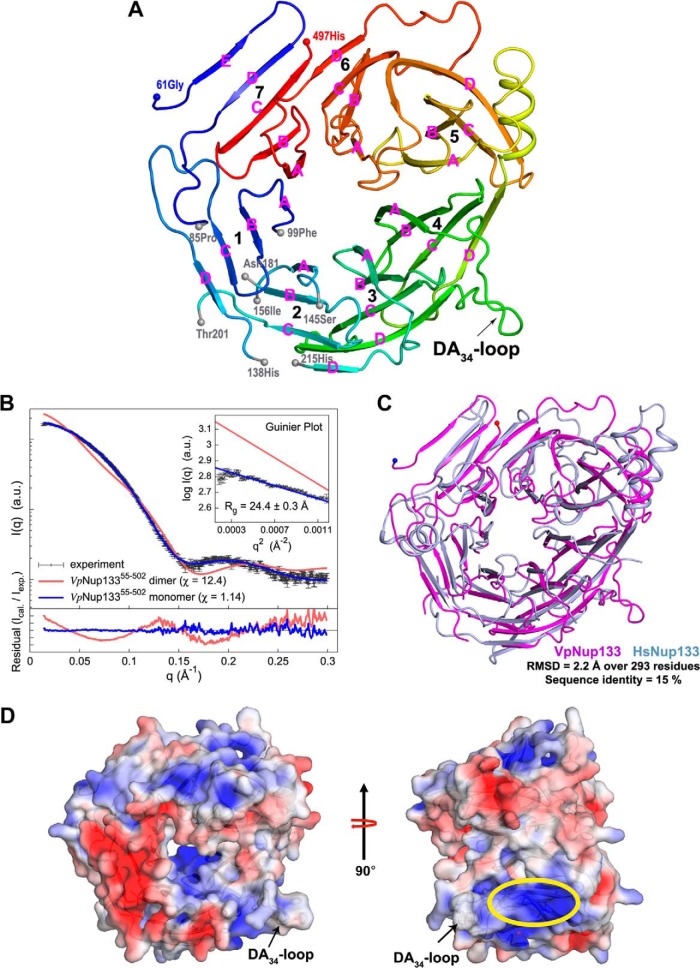
Fig. 2.
Crystal structure of VpNup13355–502. A, overall fold of VpNup13355–502 β-propeller domain is shown in cartoon representation with color shading in blue to red from the N to the C terminus. Terminal residues of the disordered segments are marked with gray spheres. The location of the DA34-loop containing a potential ALPS motif is indicated by an arrow. Secondary structure elements are shown as defined by the DSSP (86) program. Strands within each propeller blade are marked in pink. B, comparison of the merged experimental SAXS profile (black) of VpNup13355–502 with the calculated SAXS profiles from the complete dimer model (χ = 12.4, red) and the complete monomer model (χ = 1.14, blue). The lower plot presents the residuals (calculated intensity/experimental intensity) of each calculated SAXS profile. The upper inset shows the SAXS profiles in the Guinier plot with an Rg fit of 24.4 ± 0.3 Å. The maximum particle size (Dmax) was 77.8 Å (determined experimentally). C, superposition of VpNup13355–502 (shown as a pink cartoon) and HsNup13375–477 (PDB: 1XKS; shown as a light-blue cartoon) (19) structures performed using the SSM (87) routine as implemented in COOT (34). D, electrostatic potential of VpNup13355–502 plotted onto its solvent accessible surface. Missing side chains and charges were assigned for the VpNup13355–502 structure on the PDB2PQR Web server (88), and the electrostatic surface was calculated using APBS (89) within PyMOL. Negative (−4 kT/e) and positive (+4 kT/e) potentials are shown in red and blue, respectively. The positively charged surface located adjacent to the DA34-loop (annotated as the potential ALPS motif) is marked by an ellipse in yellow. Phenylalanines at positions 275 and 276 are shown in sticks as a reference for the location of the DA34-loop.