Figure 1.
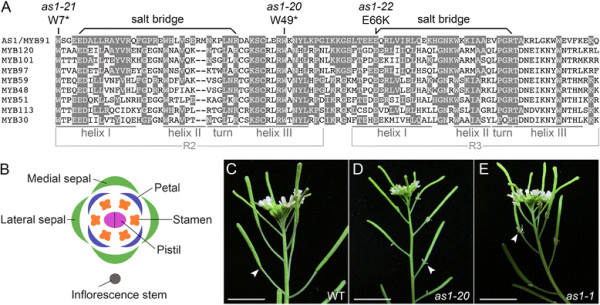
Mutations inAS1delay sepal abscission. A: Sequence alignment of the DNA-binding domain of AS1 and related Arabidopsis R2R3 MYB proteins. Conserved amino acid residues are shaded. The pairs of acidic glutamic acid (E) and basic arginine (R) residues that form salt bridges in the R2 and R3 repeats are linked by bars. The positions of three as1 mutations that alter organ abscission are indicated. The as1-20 (reference allele for this study) and as1-21 mutations introduce stop codons in place of codons specifying tryptophan. The as1-22 mutation replaces a conserved glutamic acid with a lysine; the affected residue is an integral part of the salt bridge formed in the R3 repeat of AS1. B: Diagram of a wild-type flower illustrating the orientation of the medial and lateral sepals with respect to the inflorescence stem. C: Wild-type inflorescence. Flowers shed their outer organs (stage 16), leaving the developing fruit behind. Scale bar, 1 cm. D/E: as1-20(D) and as1-1(E) mutant inflorescences. Mutant flowers typically retain their medial sepals until after the fruit is fully elongated (stage 17). Scale bars, 1 cm.
