Figure 3.
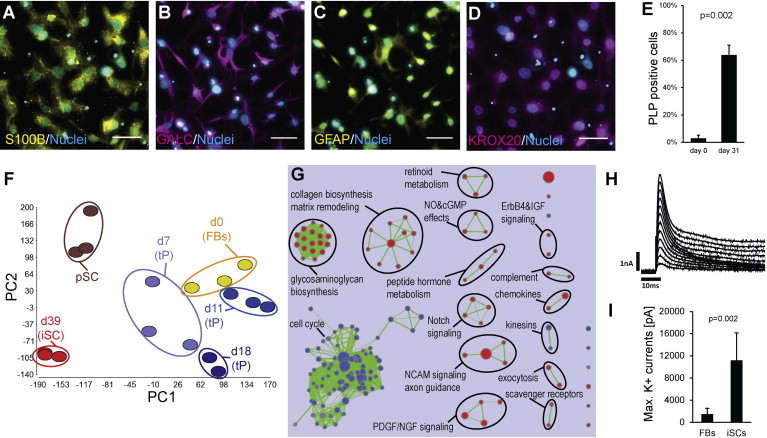
Differentiation of Transient Precursors into Induced Schwann Cells
(A–D) iSCs express Schwann cell markers. Scale bars, 50 μm.
(E) Quantification of PLP-positive cells at d31. Few PLP-positive fibroblasts are due to background signal (n = 3).
(F) Principal component analysis of whole-transcriptome expression profiles from fibroblasts (d0), transdifferentiated cells at d7 (early tP), d11 (early tP), d18 (late tP), d39 (iSCs), and primary Schwann cells (pSCs). Principal component 1 (x axis) accounts for 27.4% and principal component 2 (y axis) accounts for 16.5% of the variation of the data set. Each stage is represented by at least two data points derived from independent experiments. The clustered transcriptomic profiles at day 39 suggest the robustness of the protocol.
(G) Enrichment map of gene sets for cellular signaling pathways (Reactome/NCI Nature PID) derived from GSEA comparing iSCs (d39) with fibroblasts (d0). Red nodes represent gene sets enriched in iSCs, whereas blue nodes represent gene sets enriched in fibroblasts. Nodes are grouped and annotated by their similarity according to related gene sets. Cluster of functionally related nodes were summarized and annotated using WordCloud (n = 3).
(H) Whole patch-clamp analysis of iSCs. Voltage-dependent current obtained from a –70 to +40 mV in 10 mV increasing steps protocol from a holding potential Vh = −80 mV. Absence of early inward current confirms the deficiency of voltage-dependent Na+ channels, whereas the outward component is consistent with the presence of voltage-dependent K+ channels.
(I) Maximal voltage-dependent K+ currents are significantly higher in iSCs than in fibroblasts. Columns show means ± SD of different cells (FBs: n = 12; iSCs: n = 7) measured in two independent experiments.
