Figure 5.
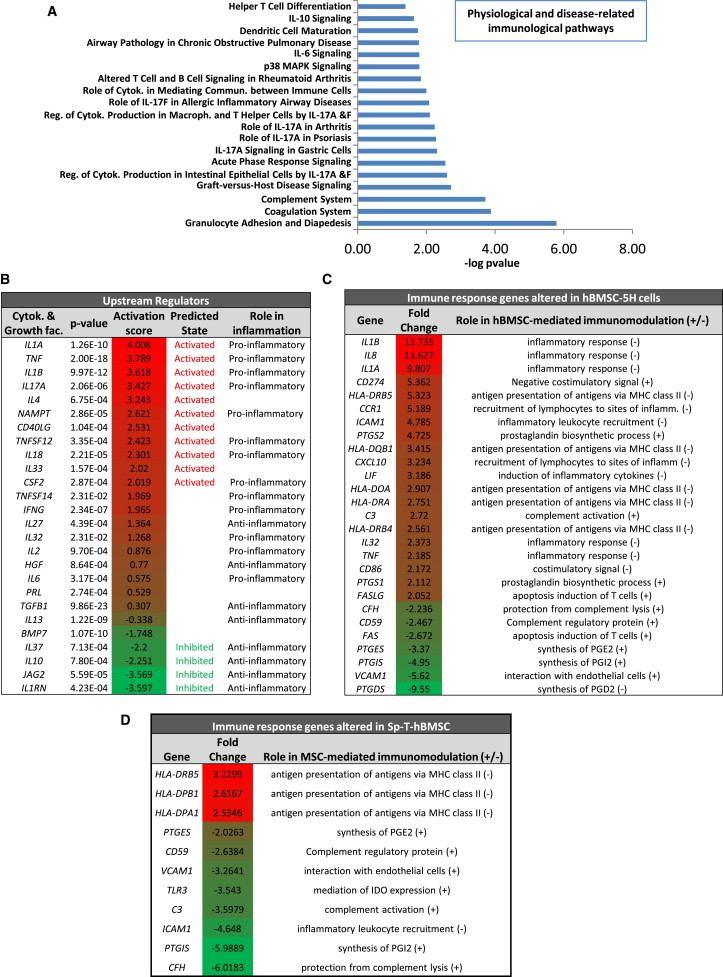
Gene-Expression Profiling Revealed an Impaired Immune Response Signaling in Transformed hBMSCs
Genes differentially expressed (p value < 0.05; regulation ≥ 2-fold) in hBMSC-5H versus hBMSC-0H were analyzed using the IPA software.
(A) Representation of the most significantly altered immunological pathways in hBMSC-5H.
(B) List of the cytokine and growth-factor-signaling pathways most significantly altered in hBMSC-5H cells. The corresponding activation Z scores are indicated and highlighted in red (activation)/green (inhibition) color scale. A regulator-signaling pathway is predicted to be active or inhibited if their Z score is higher than 2 or lower than −2, respectively. Whether each regulator functions as pro- or anti-inflammatory factor is indicated.
(C) List of genes involved in the induction (+) or inhibition (−) of immunosuppression altered in MSC-5H cells. The fold change expression is highlighted using red (activation)/green (inhibition) color scale.
(D) List of genes involved in the induction (+) or inhibition (−) of immunosuppression altered in Sp-T-hBMSC-1 as compared to syngeneic nontransformed hBMSCs. The fold change expression is highlighted using red (activation)/green (inhibition) color scale.
