Figure 3.
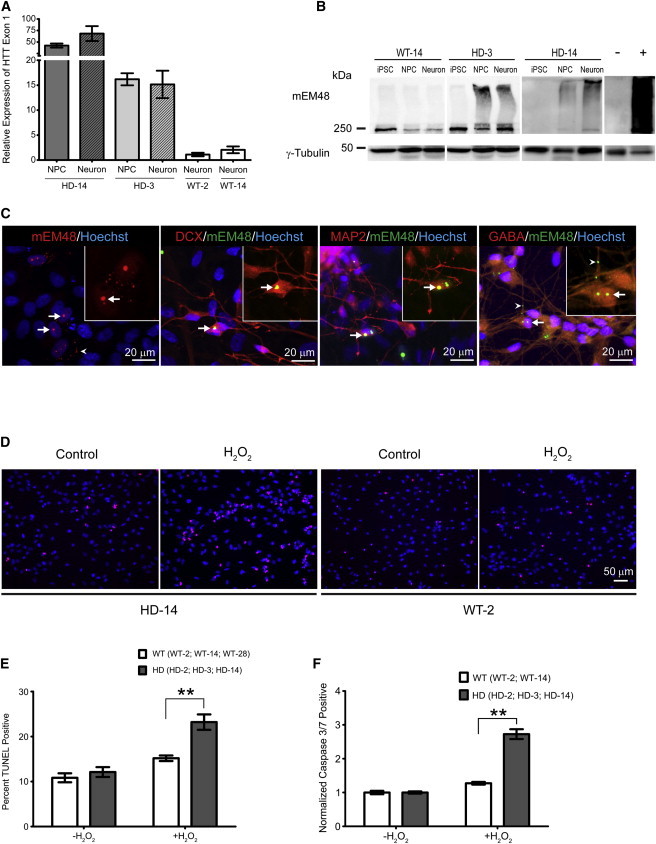
Elevated Expression and Aggregation of mHTT Accompany Increased Vulnerability to Oxidative Stress in HD Cells
(A) qRT-PCR analysis of HTT exon 1 during in vitro neural differentiation. Expression levels are compared to WT-2 neuron control line. Results were from three biological replicates. qRT-PCR samples run in duplicate. Data and error bars are represented as mean ± SEM.
(B) Western blot analysis revealed an increase in oligomeric mHTT that correlates with neuronal differentiation stage. Notice aggregate accumulation in the stacking gel (above 250 kDa). Negative (−) and positive (+) lanes are frontal cortex samples from nontransgenic and HD monkeys, respectively.
(C) Left: staining of in vitro differentiated HD-NPCs with mEM48 (red) reveals distinct intranuclear inclusions and cytosolic aggregates (arrowhead) of mHTT. Middle left: mHTT inclusions (green) colabeled with DCX (red). Middle right: mHTT inclusions (green) colabeled with MAP2 (right; red). Right: mHTT inclusions and neuropil aggregates (arrowhead; green) colabeled with GABA (red). Staining using Hoechst (blue). Arrows indicate intranuclear inclusions.
(D) Images represent TUNEL staining of WT-2 and HD-14 NPCs after 24 hr H2O2 challenge. H2O2 TUNEL-positive cells (red). Nuclear staining is shown using Hoechst (blue).
(E) Quantification of TUNEL assay on HD- and WT-NPCs following H2O2 exposure. TUNEL-positive cell (percentage) was averaged for three HD lines (HD-2, HD-3, HD-14) and three WT lines (WT-2, WT-14, WT-28). Percentage of TUNEL-positive cells was calculated from total number of cells from three biological replicates of each cell line. Data are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
(F) Quantification of caspase 3/7-positive HD-NPCs compared to WT-NPCs following H2O2 exposure. Apoptotic population percentage was averaged for three HD lines (HD-2, HD-3, HD-14) and two WT lines (WT-2, WT-14). Values represent fold change from untreated sample. Results are shown from three biological replicates of each cell line. Data and error bars are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
