Figure 4.
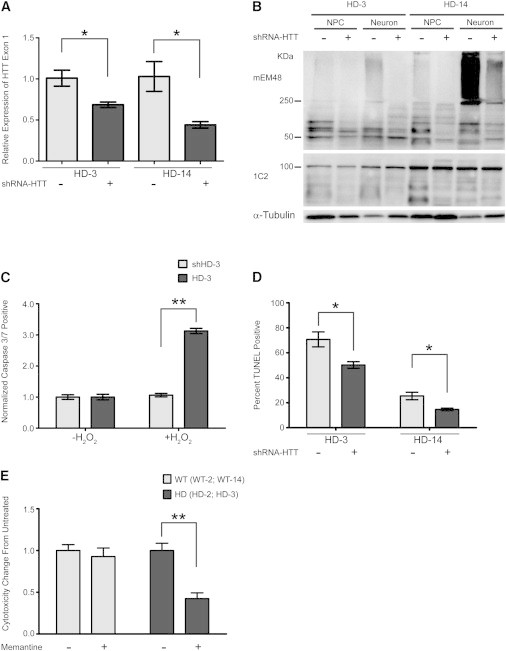
Reversal of HD-Associated Phenotypes Using shRNA and Memantine
(A) qRT-PCR analysis of HTT exon 1 expression in HD-3 and HD-14 NPCs after infection with lentivirus expressing shRNA-HTT. Results shown from three biological replicates, qRT-PCR samples run in duplicate. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
(B) Western blot analysis of mHTT aggregation in HD-3 and HD-14 NPCs and in vitro differentiated neural cells with (+) or without (−) expressing shRNA-HTT.
(C) Quantification of caspase 3/7-positive HD-3 and shHD-3 NPCs following H2O2 exposure. Values represent fold change from untreated sample. Results are shown from three biological replicates. Data and error bars are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
(D) Quantification of TUNEL assay on HD-3 and HD-14 neurons after in vitro differentiation with or without expressing shRNA-HTT. Percentage of TUNEL-positive cells calculated from total number of cells identified by Hoechst staining. Results are shown from three biological replicates. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
(E) Cytotoxicity profile of neurons treated with 10 μM memantine drug measured by G6PD assay. Values represent mean fold change in cytotoxicity from untreated cells. Cytotoxicity values averaged from two HD lines (HD-2, HD-3) and two WT lines (WT-2, WT-28) from three repeated experiments. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
