Abstract
Background
Despite the intensive use of Bacillus thuringiensis israelensis (Bti) toxins for mosquito control, little is known about the long term effect of exposure to this cocktail of toxins on target mosquito populations. In contrast to the many cases of resistance to Bacillus thuringiensis Cry toxins observed in other insects, there is no evidence so far for Bti resistance evolution in field mosquito populations. High fitness costs measured in a Bti selected mosquito laboratory strain suggest that evolving resistance to Bti is costly. The aim of the present study was to identify transcription level and polymorphism variations associated with resistance to Bti toxins in the dengue vector Aedes aegypti. We used RNA sequencing (RNA-seq) for comparing a laboratory-selected strain showing elevated resistance to Bti toxins and its parental non-selected susceptible strain. As the resistant strain displayed two marked larval development phenotypes (slow and normal), each phenotype was analyzed separately in order to evidence potential links between resistance mechanisms and mosquito life-history traits.
Results
A total of 12,458 genes were detected of which 844 were differentially transcribed between the resistant and susceptible strains. Polymorphism analysis revealed a total of 68,541 SNPs of which 12,571 SNPs exhibited more than 40% frequency difference between the resistant and susceptible strains, affecting 2,953 genes. Bti resistance is associated with changes in the transcription level of enzymes involved in detoxification and chitin metabolism. Among previously described Bti-toxin receptors, four alkaline phosphatases (ALPs) were differentially transcribed between resistant and susceptible larvae, and non-synonymous changes affected the protein sequence of one cadherin, six aminopeptidases (APNs) and four α-amylases. Other putative Cry receptors located in lipid rafts, such as flotillin and glycoside hydrolases, were under-transcribed and/or contained non-synonymous substitutions. Finally, immunity-related genes showed contrasted transcription and polymorphisms patterns between the two developmental resistant phenotypes, suggesting the existence of trade-offs between Bti-resistance, life-history traits and immunity.
Conclusions
The present study is the first to analyze the whole transcriptome of Bti-resistant mosquitoes by RNA-seq, shedding light on the importance of studying both transcription levels and sequence polymorphism variations to get a comprehensive view of insecticide resistance.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-926) contains supplementary material, which is available to authorized users.
Keywords: RNA-seq, Bacillus thuringiensis israelensis toxins, Mosquito, Bio-insecticide resistance, Toxin receptors, Lipid rafts, Resistance costs, Evolutionary trade-offs
Background
Biological control strategies involving Bacillus thuringiensis israelensis (Bti) are increasingly used as alternatives to chemicals in order to limit the spreading of mosquito-borne pathogens transmission and nuisance. During sporulation, Bti produces a composite crystalline inclusion composed of four main toxins: Cry4Aa, Cry4Ba, Cry11Aa and Cyt1Aa. Upon larval ingestion, the crystalline inclusions dissolve in the alkaline pH of the insect’s gut, releasing Cry and Cyt protoxins that will be processed to active toxins by insect gut proteases [1]. Cyt and Cry toxins are known to act in synergy to kill mosquito larvae, but the precise mechanisms of their interaction are not fully understood [2, 3]. Other Bacillus thuringiensis subspecies produce Cry toxins that are specific to their target organism, and most research has been done on Cry toxins mode of action in various insects (mostly in lepidopterans and coleopterans, but also in dipterans) and nematodes [4, 5]. The selectivity of Cry toxins is mainly due to the interaction with specific receptors present at the surface of midgut epithelium cells of the larva. The activated Cry toxins bind to specific protein receptors, insert into the membrane and form pores, resulting in midgut disruption, bacterial infection and death of the insect. In the recent years, several models have been proposed to describe the precise mode of action of Bt Cry toxins (reviewed in [6, 7]). The binding of Cry toxins can involve interaction with multiple specific receptors, including cadherin-like proteins but also aminopeptidases N (APNs) and alkaline phosphatases (ALPs) which are mostly located in lipid rafts (reviewed in [8]). In mosquitoes, besides cadherins, APNs and ALPs, α-amylases were also shown to bind to Cry4Ba and Cry11Aa toxins [9]. As compared to what is known about the mode of action of Bt Cry toxins in lepidopterans and coleopterans, the understanding of Bti mode of action in mosquito lies far behind, mainly due to the lack of Bti-resistant mosquito populations to dissect the mechanisms of resistance. Indeed, the cocktail of toxins produced by Bti, and especially the presence of Cyt toxin, appear to hinder the evolution of resistance to Bti both in the laboratory and in the field [2]. However, recent works conducted in our group revealed that Cyt1Aa toxin was strongly affected by the organic matter found in most mosquito breeding sites, inducing a decreased efficacy and a low-level persistence of Cyt toxins as compared to Cry toxins [10, 11]. This ultimately led to the modification of toxins proportions in field-persistent Bti (Cry toxins became the most abundant toxins while it is Cyt toxin in commercial Bti), as shown in leaf litters containing persistent Bti sampled in mosquito breeding sites several months after a treatment [11]. This field-collected Bti was used to select a laboratory strain for 22 generations, resistant to Bti toxins (LiTOX strain [12]). Two phenotypes were observed in the LiTOX strain: larvae with a normal development time (5 days as the parental Bora-Bora strain) named LiTOX_N, and larvae with a slow development (about 10 days) named LiTOX_S. As compared to the susceptible parental Bora-Bora strain, LiTOX_N exhibited resistance ratio (RR50) of 30.2-fold, 13.7-fold and 6.3-fold for Cry4Aa, Cry4Ba and Cry11Aa respectively. LiTOX_S exhibited the same resistance ratio as LiTOX_N for Cry4Ba and Cry11Aa but was not resistant to Cry4Aa [12]. In addition to an altered development, the resistant LiTOX strain exhibited lower female fecundity and lower egg survival, suggesting that Bti resistance is associated with high fitness costs [13]. In the present study, we used the LiTOX strain to undertake a whole transcriptome analysis by RNA-seq of the two developmental phenotypes LiTOX_S and LiTOX_N versus the susceptible parental strain aiming at dissecting the molecular pathways to resistance to Bti Cry toxins at the whole organism level, and at understanding the trade-offs underlying Cry-toxin resistance and life history traits. Transcription level and polymorphism variations associated with each resistance phenotype were identified in regards of known and new putative mechanisms involved in Bti toxins resistance and the way they might interfere with mosquito life history traits is discussed.
Results
Mapping and re-annotation results
A total of 197,175,220 short reads (75 bp) were sequenced, with an average of 33 million reads per sample. An average mapping rate of ~83% (164.66 million mapped reads) was reached. After filtering on base quality (150.86 million reads left) and on mapping quality, 129.32 million reads were retained for further analysis (Additional file 1: Table S1). A total of 12,942 transcripts showed a coverage >0.5 Reads Per Kilobase exon Model (RPKM) in at least one strain, including 2,717 transcripts showing a different structure as compared to genome annotation (21.8%) and 484 novel transcription events (NTEs) (3.7%) predicted based on spliced reads and distance from known transcripts (close or far, respectively). NTEs were not considered in further analyses. Because only 6,996 detected transcripts were functionally annotated in VectorBase (genome version AaegL2.1, vectorbase.org) we performed a Blast2GO analysis of all predicted peptides annotated as ‘conserved hypothetical protein’ or ‘hypothetical protein’ in Vectorbase against the protein database Swissprot (Blastp, E-value < 10-3, annotation cut-off = 55, GO weight = 5). This allowed re-annotation of 2,608 transcripts (Additional file 2: Table S2). We also re-annotated known mosquito Bti-toxin receptors based on bibliography [14, 15], ending up with about 77% of annotated transcripts in our dataset, including 80 putative Cry receptors (15 α-amylases, 14 ALPs, 12 cadherins and 39 APNs), and 40 trypsins/chymotrypsins potentially involved in Cry protoxin activation, and thereafter called ‘Bti candidates’. All the other transcripts were assigned to nine categories based on their annotation as follows: ‘Detoxification’, ‘Other enzymes’, ‘Cuticle’, ‘Immunity’, ‘Hormones and neurotransmitters signaling’, ‘Transcription factors’, ‘Intra or extracellular trafficking/chaperonins’, ‘Structure’ and ‘Unknown’.
Differential expression
Among all detected genes, 844 (6.8%) were differentially transcribed (Table 1) in at least one LiTOX phenotype as compared to the parental susceptible strain, including 12 candidate genes (Additional file 3: Table S3). The two LiTOX phenotypes shared 266 differentially transcribed genes as compared to the susceptible (191 and 75 genes under and over-transcribed, respectively, Figure 1), and 84 genes were over-transcribed in LiTOX_N and under-transcribed in LiTOX_S. Among genes under-transcribed in both phenotypes, GO terms associated with detoxification process were over-represented, while those associated with cuticle/chitin metabolism were over-represented in genes over-transcribed (Figure 1, Additional file 4: Table S4). In genes specifically under-transcribed in LiTOX_N, GO terms associated with immunity and ALP activity were over-represented. In LiTOX_S, GO terms associated with lipid metabolism were over-represented among the under-transcribed genes and those associated with proteolytic activity were over-represented in over-transcribed genes. Among candidate genes, some specific proteases such as the chymotrypsin AAEL015105 or the trypsin AAEL006376, potentially involved in Cry protoxin activation, were under-transcribed in both LiTOX phenotypes (Figure 2). Among genes differentially transcribed between the two LiTOX phenotypes, one serine protease (AAEL010139) was below the detection threshold in LiTOX_N (0.2 RPKM, as compared to 7.4 and 8.2 RPKM in Bora-Bora and LiTOX_S strains respectively), and five putative Cry receptors were differentially transcribed: one α-amylase (AAEL010540) and four ALPs (AAEL003286, AAEL003309, AAEL009077 and AAEL000931). Genes involved in immunity showed contrasted expression patterns in the two LiTOX phenotypes: several genes involved in the melanization process (prophenoloxidases) were under-transcribed in both LiTOX phenotypes, three defensins (antimicrobial peptides) were under-transcribed only in LiTOX_N, and several genes involved in the Toll pathway (e.g., spaetzle-like cytokine) were over-transcribed in LiTOX_N and under-transcribed in LiTOX_S. Finally, genes involved in cuticle/chitin metabolism were mostly over-transcribed in LiTOX_N and under-transcribed in LiTOX_S. Conserved domain search (CDS) revealed that all but four were chitin-binding proteins containing the typical Rebers & Riddiford (RR) consensus motif [16]. Based on phylogenetic analysis and sequences alignment, 26 (59%) contained the diagnostic motif GFxAxV (degenerated in three of them) of RR-2 cuticle proteins while 18 (41%) were RR-1 cuticle proteins [17, 18] (Figure 3).
Table 1.
Differential transcription analysis overview: Number (and proportion) of transcripts significantly over or under-transcribed in each LiTOX phenotype and altogether (LiTOX-N or LiTOX-S) as compared to the control parental strain
| LiTOX-N | LiTOX-S | Any strains | ||||
|---|---|---|---|---|---|---|
| Transcripts | % | Transcripts | % | Transcripts | % | |
| AC* test P value and FC > =2 | 564 | 4.4% | 745 | 5.8% | 923 | 7.1% |
| Over transcribed | 274 | 2.1% | 166 | 1.3% | 356 | 2.8% |
| Known transcripts | 248 | 1.9% | 150 | 1.2% | 323 | 2.5% |
| Novel transcription event | 26 | 0.2% | 16 | 0.1% | 33 | 0.3% |
| Under transcribed | 290 | 2.2% | 579 | 4.5% | 661 | 5.1% |
| Known transcripts | 269 | 2.1% | 528 | 4.1% | 605 | 4.7% |
| Novel transcription event | 21 | 0.2% | 51 | 0.4% | 56 | 0.4% |
*Audic-Clavery test.
Figure 1.
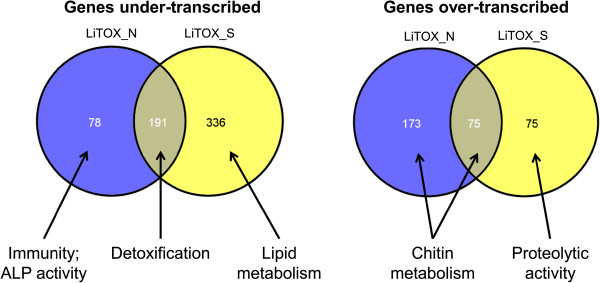
Genes differentially expressed in LiTOX phenotypes. For each Venn diagram section, the numbers of genes differentially expressed in each LiTOX phenotype as compared to the susceptible strain are indicated, together with the function(s) of these genes based on the list of GO terms significantly enriched (see Additional file 4: Table S4).
Figure 2.
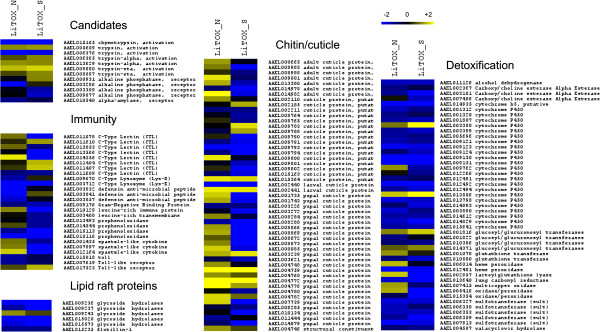
Representation of genes significantly differentially transcribed in at least one LiTOX phenotype as compared to the susceptible strain for five functions: ‘Candidates’, ‘Lipid raft’, ‘Immunity’, ‘Cuticle’ and ‘Detoxification’. The color scale corresponds to log2 transcription ratio as compared to the susceptible strain. Significant values in Additional file 3: Table S3.
Figure 3.
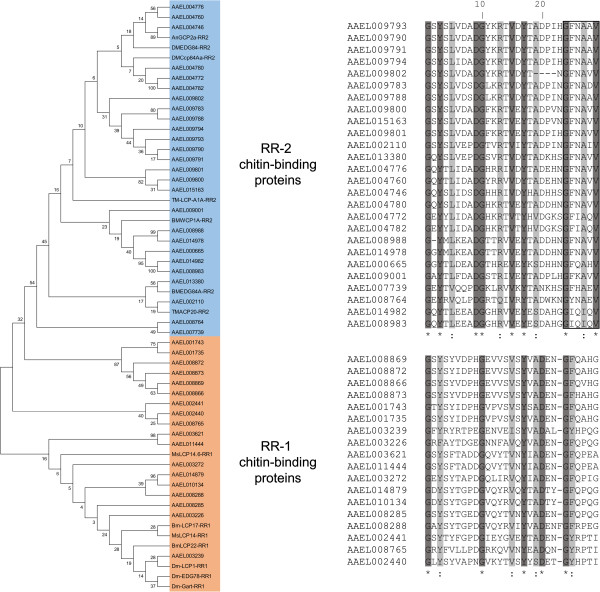
Phylogenetic analysis and sequence alignment of R&R consensus domain from the 44 differentially transcribed genes encoding chitin-binding proteins. Representatives from the two RR sub-groups from other insect species were added to support the tree (Anopheles gambiae: AnGCP2a-RR2, AAC05656; Bombyx mori: Bm-LCP17-RR1, FAA00504; BmLCP22-RR1, NP_001036828; BMEDG84A-RR2, BAA33195; BMWCP1A-RR2, BAB32475; Drosophila melanogaster: Dm-LCP1-RR1, NP_476619; Dm-EDG78-RR1, NP_524198; Dm-Gart-RR1, NP_476673; DMCcp84Aa-RR2, AAD19803; DMEDG84-RR2, P27780; Manduca sexta: MsLCP14-RR1, AAA29319; MsLCP14.6-RR1, Q94984; Tenebrio molitor: TM-LCP-A1A-RR2, P80681; TMACP20-RR2, P26967. Bootstrap values (2000 replicates) are shown on each branch of the tree. Symbols below the aligned amino acids indicate identity (*), or high conservation (:). Identical and highly conserved amino acids are highlighted in dark grey and light grey, respectively. The GFxAxV motif, diagnostic of the RR-2 sub-group, is highlighted by a box.
SNPs detection and analysis
A total of 166,943 SNPs were called, of which 68,541 had a total coverage ≥30 and affected 6,511 genes distributed over ~67% (1106/1636) of the Ae. aegypti supercontigs that have annotated genes, including 66 candidate genes (50 receptors and 16 trypsins/chymotrypsins). A total of 2,953 genes were affected by 12,571 differential SNPs (more than 40% allelic frequency difference between the susceptible and resistant strains). Little overlap was found between genes differentially transcribed in selected strains and those affected by differential SNPs: only 76 differentially expressed genes were affected by non-synonymous differential SNP (Additional file 3: Table S3). This was expected as RNAseq data are restricted to transcripts and did not cover regulatory regions often located outside transcript boundaries. While in genes differentially transcribed, three categories of genes were over-represented (‘chitin/cuticle’, ‘detoxification’ and ‘immunity’), two other categories of genes (‘intra/extra cellular trafficking’ and ‘enzymes’) were over-represented in genes affected by differential SNPs between the resistant and susceptible strains (Figure 4). Thirty two candidates (25 putative toxin receptors and 7 trypsins) were affected by a total of 508 SNPs from which 133 (26.3%) were differential SNPs; although this is not significantly different from the proportion of differential SNPs in all other genes affected, non-synonymous mutations were significantly more abundant in candidates than in any other affected genes (Fisher exact test, p = 10-4; Figure 5). Among the putative Cry receptors, one cadherin, 12 aminopeptidases and 8 α-amylases were affected by differential SNPs (Additional file 5: Table S5). This included 27 non-synonymous SNPs affecting the protein sequence of 1 cadherin, 8 APNs and 4 α-amylases (Table 2). There was little difference in sequence between the two resistant phenotypes, with only 330 differential SNPs (including 14 non-synonymous changes) affecting 130 genes, none of them being candidate genes (Additional file 6: Table S6).
Figure 4.
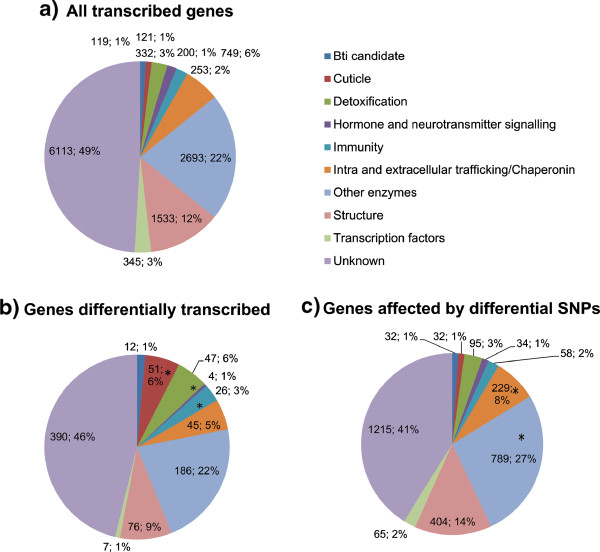
Biological functions of a) all detected genes (12,458), b) of genes differentially transcribed (844 genes) and c) of genes affected by differential SNPs between the LiTOX and the susceptible strains (2,953 genes). Genes were categorized into 10 categories: Categories were defined as follow: ‘Candidate’, ‘Detoxification’, ‘Other enzymes’, ‘Cuticle’, ‘Immunity’, ‘Hormones and neurotransmitters signaling’, ‘Transcription factors’, ‘Intra or extracellular trafficking/chaperonins’, ‘Structure’ and ‘Unknown’. Enrichment of these categories compared to all detected transcripts was then computed using a one-side Fisher’s exact test followed by Benjamini and Hochberg multiple test correction. Categories showing a corrected P value <0.05 are indicated with a star.
Figure 5.
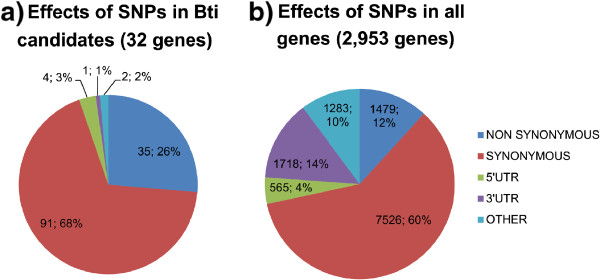
Distribution of the effects of differential SNPs a) in the 32 candidate genes containing differential SNPs and b) in all 2,953 genes containing differential SNPs. SNP effects were categorized into ‘non-synonymous’, ‘synonymous’, ‘3’UTR’, 5’UTR’, or ‘other’.
Table 2.
Non-synonymous changes in putative Cry toxin receptors, with their frequency and coverage in the susceptible and resistant phenotypes
| Bora | LiTOX_N | LiTOX_S | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Receptor | Gene ID | Reference | Variant | % | Coverage | % | Coverage | % | Coverage | Position cDNA | Amino acid position | Amino acid change | Toxin 1 |
| APN1 | AAEL009108 | C | A | 4.1 | 145 | 55.1 | 98 | 51.9 | 106 | 2539 | 847 | A- > S | |
| APN2 | AAEL003227 | C | T | 26.6 | 30 | 63.6 | 11 | 88.9 | 9 | 2240 | 747 | T- > I | |
| APN2 | AAEL005808 | C | A | 30.4 | 46 | 73.3 | 45 | 91.4 | 70 | 2289 | 763 | M- > I | Cry4Ba [40] |
| AAEL005808 | C | T | 31.4 | 51 | 85.7 | 91 | 95.5 | 110 | 2725 | 909 | V- > I | Cry4Ba [40] | |
| AAEL005808 | C | T | 15.5 | 103 | 77.7 | 130 | 75.7 | 107 | 1762 | 588 | A- > T | Cry4Ba [40] | |
| AAEL005808 | T | G | 14.5 | 117 | 84.7 | 111 | 89.4 | 132 | 1145 | 382 | E- > A | Cry4Ba [40] | |
| APN2 | AAEL005821 | A | G | 5.5 | 255 | 50.0 | 82 | 63.3 | 150 | 1660 | 554 | T- > A | |
| AAEL005821 | T | C | 5.0 | 260 | 56.1 | 98 | 65.0 | 183 | 1646 | 549 | M- > T | ||
| AAEL005821 | G | A | 10.1 | 179 | 71.6 | 88 | 78.5 | 158 | 1459 | 487 | D- > N | ||
| APN4 | AAEL007204 | C | T | 20.2 | 263 | 67.4 | 307 | 70.7 | 314 | 167 | 56 | T- > M | |
| AAEL007204 | G | C | 40.0 | 190 | 93.2 | 294 | 95.8 | 239 | 216 | 72 | E- > D | ||
| AAEL007204 | C | T | 33.8 | 142 | 93.9 | 197 | 92.7 | 220 | 283 | 95 | H- > Y | ||
| APN4 | AAEL012774 | C | T | 43.4 | 459 | 88.3 | 677 | 86.9 | 540 | 311 | 99 | G- > R | Cry11Aa [46] |
| APN5 | AAEL002696 | A | G | 23.1 | 65 | 56.3 | 48 | 64.6 | 65 | 472 | 158 | K- > E | |
| AAEL002696 | G | A | 57.7 | 71 | 100.0 | 66 | 100.0 | 59 | 2380 | 794 | V- > M | ||
| AAEL002696 | C | A | 17.8 | 90 | 66.7 | 42 | 60.4 | 48 | 2432 | 811 | A- > E | ||
| AAEL002696 | G | A | 12.3 | 57 | 70.5 | 44 | 70.0 | 50 | 746 | 249 | R- > K | ||
| APN5 | AAEL003666 | A | G | 24.6 | 126 | 63.9 | 72 | 70.8 | 106 | 1814 | 605 | V- > A | |
| AAEL003666 | C | A | 16.9 | 83 | 64.9 | 74 | 73.0 | 89 | 595 | 199 | A- > S | ||
| APN7 | AAEL007201 | A | G | 25.2 | 163 | 76.1 | 46 | 96.4 | 28 | 2219 | 740 | N- > S | |
| Cadherin | AAEL018140 | T | A | 12.1 | 33 | 43.9 | 57 | 58.7 | 63 | 790 | 160 | H- > L | Cry11Aa [15] |
| α-amylase | AAEL000642 | A | T | 24.5 | 49 | 67.9 | 56 | 47.0 | 66 | 1424 | 468 | T- > S | |
| α-amylase | AAEL007673 | C | T | 18.0 | 589 | 51.3 | 493 | 75.0 | 559 | 47 | 16 | S- > N | |
| α-amylase | AAEL010537 | AA | TT | 12.2 | 395 | 49.7 | 447 | 55.4 | 482 | 1706 | 564 | I- > F | |
| AAEL010537 | A | G | 15.6 | 825 | 53.0 | 1078 | 58.7 | 1368 | 1324 | 437 | K- > R | ||
| AAEL010537 | C | T | 16.0 | 268 | 60.5 | 248 | 64.3 | 412 | 18 | 2 | R- > W | ||
| AAEL010537 | C | T | 21.7 | 1154 | 76.1 | 804 | 83.0 | 833 | 1552 | 513 | A- > V | ||
| α-amylase | AAEL014710 | G | A | 9.9 | 81 | 60.0 | 35 | 56.5 | 23 | 992 | 331 | A- > V | |
1Toxin shown to bind this receptor in the literature.
Discussion
Two main mechanisms of resistance to Cry toxins have been described in insects so far: altered protoxin activation by gut proteases, and modification in transcription level and/or protein sequence of Cry receptors resulting in lower or failure in toxin binding [19–21]. The study of resistance to Cry toxins in insects has mostly been based on the study of a few candidate proteins first identified in lepidopterans, either toxin receptors or proteases, using proteomic approaches (2D-DIGE, ligand blotting, etc.). As compared to other Bacillus thuringiensis subspecies that typically produce a single Cry toxin, Bti produces a mixture of four different toxins, and resistance to Bti is likely to involve multiple distinct mechanisms, which may affect the physiology and metabolism of resistant insects. Altered metabolism in Bti resistant mosquitoes was revealed by fitness costs expressed at any mosquito life-stage (larval development time, fecundity, egg survival) [13]. These costs can result from pleiotropic effects of resistant alleles on life-history traits (direct cost) or more indirectly from resource reallocation between different metabolic pathways.
Global changes in gene expression and metabolic trade-offs
The two LiTOX phenotypes share common global transcriptional responses: an over-transcription of genes involved in proteolytic activity, cell cytokinesis and wound healing, and an under-transcription of enzymes classically involved in the detoxification of chemicals such as chemical insecticides, plants allelochemicals and pollutants (cytochrome P450 monooxygenases, esterases, glutathion S-transferases, UDP-glycosyltransferases, etc.). Conversely, Bti toxins are large proteins and their degradation requires a proteolytic processing rather than the activity of detoxification enzymes. Therefore, in the absence of chemical selection pressure, the under-transcription of these enzymes might represent a metabolic trade-off in the LiTOX strain to compensate the increased expression of enzymes involved in protein degradation [22]. The over-transcription of several proteases (endopeptidases, serine proteases) is in accordance with an observed increase in proteolytic activities of midgut enzymes of LiTOX larvae [23]. This increased proteolytic activity in Bti resistant larvae could reflect a higher ability to degrade toxins. Despite this global trend, particular proteases such as the metalloproteinases AAEL011547, AAEL011550 and AAEL014514, and the chymotrypsin AAEL015105 or the trypsin AAEL006376, which are putatively involved in Cry protoxin activation, were under-transcribed in both LiTOX phenotypes. The lack of metalloproteinase activity in the LiTOX strain was evidenced by the analysis of enzymatic activities in the midgut of larvae using various specific enzyme inhibitors [22]. Down-regulation of specific proteases might result in a lower and/or improper toxin activation which could also confer resistance. In lepidopterans, analysis of proteinase activities from gut extracts have shown the lack of trypsin-like and of chymotrypsin-like activity in Bt-resistant strains [20]. Furthermore, the role of proteases in resistance to Bt toxin Cry1Ca1 was demonstrated in the caterpillar Spodoptera frugiperda where a serine-protease gene under-transcribed in the midgut of intoxicated larvae was further shown by RNAi-mediated knockdown to be involved in both reduced protoxin activation in the midgut and reduced susceptibility of caterpillars to toxins in bioassays [24].
Lipid rafts and Bti-toxins trafficking through the gut membrane
Lipid rafts are membrane domains enriched in glycosylphosphatidylinositol (GPI)-anchored proteins that have been implicated in transmembrane protein trafficking and were shown to play a role as portals of entry for many pathogens including viruses, bacteria and their toxins [25, 26]. Several studies suggested that lipid rafts might play a central role in Bt-toxins toxicity. For example, the presence of lipid rafts was shown to be essential for the toxicity of Cry1C on Sf9 Lepidoptera cells [27]. However, the specific proteins associated with this mechanism have not been identified yet. A flotillin, a protein found in lipid rafts and described as a toxin transporter [28], was under-transcribed in both LiTOX phenotypes. Flotillin was shown to co-localize with Cry4Ba toxin in Aedes aegypti larval gut brush border membranes [25], and further investigation of its Cry-binding ability will contribute to characterize its role in Bti toxin resistance. Four glycoside-hydrolases were under-transcribed in both LiTOX phenotypes. Although never described as Cry toxin receptors, glycoside-hydrolases were shown to be associated with lipid rafts in the gut membrane of Aedes larvae [25]. Lipid rafts appear to play a central role in pore-forming toxins insertion in the gut membrane by functioning as platforms to recruit GPI-anchored proteins including APNs, ALPs, and glycoside-hydrolases. The role of some GPI-anchored proteins (flotillin and glycoside-hydrolases) largely present in lipid rafts of Aedes larvae guts has been so far underexplored in studies aiming at understanding Cry toxins toxicity in insects, and the present study pinpoints them as new potential candidates in Aedes.
Differences between the two LiTOX phenotypes
Several gene families were differentially transcribed between the two developmental phenotypes: genes involved in cuticle/chitin metabolism, prophenoloxidases, and genes coding for enzymes involved in lipid metabolism.
In our dataset, the category of cuticle/chitin proteins contained the highest number of genes differentially transcribed (Figures 2 and 4b, Additional file 3: Table S3). Cuticle is an exoskeleton constituted of chitin that provides physical support and protects the insects from physical injuries, desiccation and infections. Cuticle is typically divided into hard cuticle, generally sclerotized and mechanically stiff, and soft cuticle, a more flexible region that allows locomotion and appendix movements [29]. Chitin is always associated with chitin-binding proteins; although their exact function is to be investigated, it has been hypothesized that they could play important roles in cuticle metabolism and structure preservation [30]. Nearly all the genes identified as differentially transcribed in this category contained the R&R chitin-binding motif and were associated with the soft cuticle (RR-1 cuticle proteins) or with the hard cuticle (RR-2) [17]. The global under-transcription of these genes in the LiTOX_S could partly explain the slow development of this cohort, considering that a low chitin metabolism is believed to slow-down the molting process and increase the development duration. Nevertheless, it is intriguing why only genes encoding R&R-containing proteins were found differentially transcribed while genes encoding other chitin-binding proteins with peritrophin-A chitin-binding domains (ChtBD2 domain) and chitin metabolism enzymes (such as chitinases) were not differentially expressed. Also, it is challenging to know if this pattern is a side-effect consequence of the resistance phenotype or if it is part of the mechanisms of resistance developed by the insects against Cry toxins. A better understanding of the role of such proteins in the cuticle metabolism is mandatory to help deciphering their potential involvement in resistance phenotypes in insects.
Prophenoloxidases are involved in the melanization process, and as such they have a dual role: they contribute to cuticle formation (and larval development), and they participate in the immune response against pathogens by their encapsulation [31]. In Ae. aegypti, both defensins and phenoloxidases were shown to participate to immune response against bacterial challenge [32]. In our dataset, prophenoloxidases were under-transcribed in both LiTOX phenotypes, while defensins were under-transcribed only in LiTOX_N (Figure 2, Additional file 3: Table S3). Furthermore, a correlation was observed between age at pupation (development speed) and mosquito's ability to melanize Sephadex beads (immuno-capacity), suggesting that an increased immunity is costly and results in a slower development [33]. The two developmental phenotypes observed in the resistant mosquito strain LiTOX might be the result of a trade-off between immunity and development.
When focusing on candidate genes, only few differences in transcription level and no significant difference in polymorphism were observed between the two LiTOX phenotypes (e.g. between LiTOX_N and LiTOX_S). The serine protease AAEL010139 was not expressed in LiTOX_N, the Cry4Aa-resistant cohort. Among the putative Cry-toxin receptors, four alkaline phosphatases (ALPs) were differentially transcribed between the two LiTOX phenotypes: ALP AAEL003286 was specifically under-transcribed in LiTOX_N, which may be linked to the high Cry4Aa resistance of this phenotype. Three other ALP were specifically under- (AAEL003309 and AAEL009077) or over-transcribed (AAEL000931) in LiTOX_S. Cry4Aa mode of action and mechanisms of resistance are the least investigated and understood among the Bti toxins. Functionally validating the involvement of these few candidate genes could help lifting the veil on the specific mechanisms associated with Cry4Aa resistance and provide a better understanding of the patterns of cross-resistance between Bti Cry toxins.
Polymorphisms rather than expression changes are found in putative Bti-toxins receptors
Among the 80 putative Cry receptors detected in larvae, only 5 were differentially transcribed in the LiTOX strain as compared to the susceptible strain. Among these, the α-amylase AAEL010540 was under-transcribed in both LiTOX phenotypes sharing resistance to Cry4Ba and Cry11Aa. In An. albimanus, an α-amylase (aamy1) has been described as a Bti toxin receptor, binding both Cry4Ba and Cry11Aa [9]. Although this requires functional validation, this suggests that the Ae. aegypti α-amylase AAEL010540 (48% identity with aamy1) could bind Cry toxins. No cadherin or APN that were previously associated with Cry resistance in insects [34–37] or showed to bind Bti Cry toxins in mosquitoes [38–40] were differentially transcribed in the LiTOX strain, suggesting that if they are involved in Bti resistance in LiTOX, this is not related to an altered expression but rather to changes in protein sequence affecting their affinity for Cry toxins. Indeed, one cadherin, eight APNs and four α-amylases displayed differential SNPs leading to non-synonymous changes between the resistant and susceptible strains.
Cadherin
CAD-like proteins have been the most intensively studied putative Cry toxin receptor molecules in lepidopteran and coleopteran larvae [41–43]. It has been proposed that they act as the first receptors of Cry toxins, binding toxin monomers and facilitating further processing required for the pre-pore oligomer formation [44]. Although twelve CAD-like proteins were detected in our dataset, none was differentially transcribed, and only one (AAEL018140) contained several differential SNPs (Table 2). This cadherin was previously shown by proteomic approaches to act as a Cry11Aa toxin receptor [15]. It contains a N-terminal signal peptide, 11 cadherin repeats (CR1 to CR11), a membrane-proximal region (MPR) and a trans-membrane domain. The only differential non-synonymous SNP distinguishing Bti-resistant and susceptible strains (H160L) is located in the N-terminal region preceding the first cadherin repeat (CR1), while overlay assays and immune-blotting localized Cry11Aa and Cry4Aa toxin-binding regions in CR8-CR11 [15]. More generally, Cry-toxin binding sites were usually described in the membrane-proximal region of insect cadherins, not in the N-terminal extracellular region [8]. However, amino-acid substitution at any location can dramatically change the secondary structure of a protein; for instance, the three cadherin alleles conferring resistance to Bt in the pink bollworm Ectinophora gossypiella are not located within the Cry1Aa binding site but upstream [45]. In this case, the deletion of several amino-acids upstream the toxin binding site was shown to alter the full-length cadherin and impede toxin binding. In the H160L mutation observed at relatively high frequency in the LiTOX strain (49-58%), a polar and positively charged amino acid (Histidine) is replaced by a non-charged hydrophobic amino acid (Leucine), which can result in a change in the spatial arrangement of the cadherin in the cell membrane. Further functional studies and in-silico protein modelling could allow investigating whether this change affects cadherin conformation and Cry toxin binding affinity. Altogether, our results suggest that cadherin might not have a central role in resistance to Bti in mosquitoes.
APNs and α-amylases
APNs and α-amylases are digestive enzymes that play a key role in larval nutrition and development, being involved in protein and glucose metabolism, respectively. APNs have been extensively studied as putative Cry toxin receptors in many insects and were classified into 8 classes based on their sequence identity [8, 14]. APNs from several classes were reported to bind with more or less affinity to the different Cry toxins in various insects, including AeAPN1 in Aedes and APN2 in Anopheles [14], but their precise role in Bti toxicity remains to be elucidated. Out of the 31 APNs detected in Ae. aegypti larvae, 12 contained differential SNPs, and non-synonymous differential changes affected the sequence of 8 of them (Table 2). In the APN2 AAEL005808 recently shown to be a functional receptor of Cry4Ba toxin [40], 4 non-synonymous changes were nearly fixed in the LiTOX strain (both phenotypes), suggesting ongoing selective sweep on this gene. One non-synonymous change affected the sequence of the APN1 AAEL012778 previously shown to bind Cry11Aa [46]. Although not usually cited as a Cry-toxin receptor, an α-amylase was recently shown to bind Cry4Ba and Cry11Aa in Anopheles [9]. Our dataset pinpoints five α-amylases potentially involved in Aedes resistance to Bti toxins: one α-amylase was under-transcribed only in LiTOX_N (resistant to all Cry toxins), and four other α-amylases presented sequence changes in both cohorts of the resistant LiTOX strain as compared to the susceptible strain. Because of their key role in insect nutrition, altered expression of these digestive enzymes as a resistance mechanism is likely to be costly in terms of larval development, and this might explain why the observed changes in these enzymes are mostly amino-acid changes rather than changes in expression levels. Indeed, change in the protein sequence might dramatically alter toxin binding site while having small effects on digestive efficiency. Further studies on the differential abilities of these α-amylases to bind Bti Cry toxins and to act as functional toxin receptors are required to determine their role in Bti toxicity. Also, molecular modelling of APN is necessary to identify the oligosaccharide that are involved in the binding of Cry toxins to verify if the SNPs described in this study can alter the Cry-APN interaction and partly explain the resistance phenotype observed [47].
Conclusions
The present study is the first to analyze the whole transcriptome of Bti-resistant larvae by deep sequencing, allowing studying change in gene expression level and sequence polymorphisms using the same dataset. It reveals dramatic modifications in the transcriptional profiles selected in resistant larvae, with a global transcriptional increase of genes coding for proteases and chitin-binding proteins, and a decrease in transcription of genes involved in detoxification and immunity. Whether these modifications are directly involved in Bti-toxin processing or more indirectly through complex metabolic compensations selected to limit the cost of resistance remains to be functionally investigated. Regarding candidate Bti-toxin receptors, our dataset relativizes the role of cadherin in resistance to Bti in mosquitoes, and highlights the importance of studying changes in the sequence rather than in transcription levels of APN and ALP, but also of other proteins involved in protein trafficking through the cell membrane such as flotillins and glucoside hydrolases.
Methods
Strains and Sample preparation
The Ae. aegypti laboratory strain Bora-Bora susceptible to all insecticides was selected using field-collected leaf-litter containing Bti toxins (LiTOX strain) as described in [12]. At each generation, an average of 6,000 larvae was exposed to toxic leaf litter in order to obtain a mortality rate of 70%, so that about 1,800 adults constituted the next generation, limiting bottleneck effects in the LiTOX strain. Mosquito strains were reared in standard insectarium conditions (27°C, 14 h/10 h light/dark period, 80% relative humidity). The average generation turnover was 45 days and selection was carried out over 18 generations. Larvae were bred in the insectarium till they reached early 4th instar. LiTOX_S larvae were allowed to develop for 5 days more than other larvae. For each of the three phenotype (Bora-Bora, LiTOX_N and LiTOX_S) total mRNA was extracted from three pools of 60 4th-stage larvae (reared in different batches and originating from different egg-laying female pools) at the same time of the day, using the RNAqueous-4PCR kit (Applied Biosystems/Ambion, Austin, TX, USA), according to the manufacturer’s instructions. RNA quantity and quality was checked using bioanalyzer, and RNA from the 3 biological replicates was pooled in equal proportion to obtain one representative total RNA sample per phenotype (each made from 180 individuals). For each of the three phenotypes, two distinct cDNA libraries were prepared from the same pool of total mRNA and sent at the National Genotyping Center (Genoscope, France) and sequenced individually on the Illumina Genome Analyser II system (GAII) to assess technical variations.
Short read mapping and assembly on the reference genome
The Tophat algorithm (release 2.0.9, http://ccb.jhu.edu/software/tophat) was applied with defaults parameters to align all the reads onto the Aedes aegypti reference genome AaegL2.1 (http://ccb.jhu.edu/software/tophat) by taking into account, both already known (--no-novel-juncs) and novel ab initio splice exon-exon junctions [48].
Estimation of transcripts’ relative abundance and differential expression (DE) analysis
Bam files were then loaded into Genespring NGS Version 12 (Agilent) software. Reads were filtered on base quality (mean base quality >30 and <10 N per reads) and on mapping quality (alignment score > =97 and Mapping quality > = 250 and remove non-primary multiply mapped reads). Ae. aegypti genome was then re-annotated based on reads distribution and RPKM calculated for each known or new putative transcript using default parameters (min exon length percentile 10, min intro length percentile 10, max intron length percentile 90, min exon RPKM percentile 50, min gene RPKM percentile 50, min gene length percentile 10, min exon RPKM with respect to host gene percentage 75, minimum number of reads in exon 10). For each strain, RPKM correlation between technical replicates was calculated. As r2 were >0.9 for all strains (acceptable technical variation, see Additional file 7: Figure S1), technical replicates were pooled for further analyses, in order to reach a high coverage per transcript and per SNP position. Transcripts with >0.5 RPKM in at least one strain were retained for differential transcription analysis. Differential transcription level was tested for each transcript between each resistant phenotype (LiTOX_N and LiTOX_S) and the parental strain Bora-Bora using an Audic Clavery test designed to compare two cDNA libraries [49]. This Bayesian method is based on the assumption that under the null hypothesis, read counts of the same gene in two libraries come from the same but unknown Poisson distribution. The posterior probability is obtained by Bayesian averaging (infinite mixture) of all possible Poisson distributions with mixing proportions equal to the posteriors under the flat prior. This probability is further adjusted for multiple testing (Benjamini and Hochberg correction). Despite this popular method has been validated by the statistician community [50], we recommend sequencing biological replicates rather than pooling them prior sequencing in future RNAseq analyses, as sequencing costs are now acceptable and the later approach allows a more robust identification of genes affected by variations across conditions. Transcripts showing an adjusted P- value <10-15 and a fold change ≥2 fold in either direction were considered differentially transcribed.
GO term analysis
The annotation terms from the GO ontology were retrieved from the VectorBase-UniprotKB links. For each resistant phenotype and each differential expression state (i.e. up or down regulated as compared to the Bora-Bora strain), GO terms significantly over-represented were determined using a corrected P-value threshold of 0.05.
SNP detection and analysis
Detection of polymorphisms was performed based on the 129.32 million reads passing quality filters with the following parameters: confidence score threshold =100, coverage >20 reads, base quality cut off =5, ignore locations within or next to homopolymer stretches >10 nucleotides. Among all detected polymorphism variations, only SNP substitutions were considered for differential polymorphism analyses. SNP allele frequencies were then computed between each LiTOX phenotype and the susceptible strain. In a previous study on the LiTOX strain, we have validated the reliability of mRNA sequencing pooled data for inferring population allelic frequencies on 269 SNP: the correlation between allelic frequencies obtained from pooled mRNA sequencing (180 pooled larvae) and individual genotypes (N = 28 individuals) obtained using a DNA Illumina GoldenGate array was highly significant (P <0.001, r =0.85) [51]. Allele frequencies were considered as differential between a LiTOX phenotype and the susceptible strain (hereafter named as differential SNPs) if the following conditions were fulfilled: total read coverage at SNP position ≥30 and allelic frequency difference between both strains ≥40% in either direction. Genic effects of SNPs were computed by comparing SNP locations with reference genome annotation, and were defined as 5’UTR, synonymous, non-synonymous, 3’UTR and other (intronic or intergenic- i.e. close but not within gene boundaries).
Phylogenetic analysis and sequence alignment of R&R consensus domain from the differentially transcribed genes encoding chitin-binding proteins
Phylogenetic analysis (neighbor joining) was performed using MEGA 6.06 software on the chitin-binding domains identified using the CDS search software (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Representatives from the two RR sub-groups from other insect species were added to support the tree (Anopheles gambiae: AnGCP2a-RR2, AAC05656; Bombyx mori: Bm-LCP17-RR1, FAA00504; BmLCP22-RR1, NP_001036828; BMEDG84A-RR2, BAA33195; BMWCP1A-RR2, BAB32475; Drosophila melanogaster: Dm-LCP1-RR1, NP_476619; Dm-EDG78-RR1, NP_524198; Dm-Gart-RR1, NP_476673; DMCcp84Aa-RR2, AAD19803; DMEDG84-RR2, P27780; Manduca sexta: MsLCP14-RR1, AAA29319; MsLCP14.6-RR1, Q94984; Tenebrio molitor: TM-LCP-A1A-RR2, P80681; TMACP20-RR2, P26967). Alignments were performed using the ClustalW software from PBIL Expasy tool (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html).
Ethics
Mosquito rearing was performed according to protocol CEEA-LR-13002 agreed by the French National Committee of ethics for animal experimentation.
Data access
RNAseq data are available at EBI/SRA/ArrayExpress (accession number E-MTAB-1635).
Electronic supplementary material
Additional file 1: Table S1: Sequencing and mapping statistics. (XLSX 15 KB)
Additional file 2: Table S2: Blast2GO analysis of all predicted peptides annotated as ‘conserved hypothetical protein’ or ‘hypothetical protein’ in Vectorbase against the protein database Swissprot (Blastp, E-value < 10-3, annotation cut-off = 55, GO weight = 5). (XLSX 134 KB)
Additional file 3: Table S3: List of genes differently expressed between resistant and susceptible Bora-Bora strains classified according to their category. (XLSX 2 MB)
Additional file 4: Table S4: GO terms enrichment analysis. Analysis was performed on transcripts significantly differentially expressed in each LiTOX phenotype as compared to the susceptible strain. GO terms associated with each transcript were extracted from Vectorbase. GO terms showing adjusted P values <0.05 were considered significantly enriched. (XLSX 17 KB)
Additional file 5: Table S5: List of supercontigs and genes affected by differential SNPs and their effects. The total number of SNPs affecting these genes is also shown. (XLSX 315 KB)
Additional file 6: Table S6: List of genes with differential SNPs between LiTOX_N and LiTOX_S; non-synonymous changes are indicated. (XLSX 19 KB)
Additional file 7: Figure S1: RPKM correlations between cDNA library replicates. Each dot represents one transcript. Only transcripts showing more than 0.5 RPKM are shown. (XLSX 2 MB)
Acknowledgements
This work was funded by the Région Rhône-Alpes (CIBLE program) and the French Agence Nationale de la Recherche (ANR-08-CES-006-01 DIBBECO). We thank T. Gaude and S. Veyrenc for their technical assistance in mosquito rearing.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LD conceived the study, analyzed the data, performed the statistical analysis and wrote the manuscript. RS and MP participated in the data analysis. GT carried out the analysis of cuticle/chitin genes and helped to draft the manuscript. VN carried out the mapping of reads on the genome. AB, JPD and SR performed the RNA extraction. JPD conceived the study, and participated in its design and analysis and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Laurence Després, Email: laurence.despres@ujf-grenoble.fr.
Renaud Stalinski, Email: renaud.stalinski@gmail.com.
Guillaume Tetreau, Email: guillaume.tetreau@gmail.com.
Margot Paris, Email: margotparis1@gmail.com.
Aurélie Bonin, Email: aurelie.bonin@ujf-grenoble.fr.
Vincent Navratil, Email: vincent.navratil@prabi.fr.
Stéphane Reynaud, Email: stephane.reynaud@ujf-grenoble.fr.
Jean-Philippe David, Email: jean-philippe.david@ujf-grenoble.fr.
References
- 1.Lacey LA. Bacillus thuringiensis serovariety israelensis and Bacillus sphaericus for mosquito control. J Am Mosq Control Assoc. 2007;23(2):133–163. doi: 10.2987/8756-971X(2007)23[133:BTSIAB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Wirth MC, Park HW, Walton WE, Federici BA. Cyt1A of Bacillus thuringiensis delays evolution of resistance to Cry11A in the mosquito Culex quinquefasciatus. Appl Environ Microbiol. 2005;71(1):185–189. doi: 10.1128/AEM.71.1.185-189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez C, Fernandez LE, Sun JG, Folch JL, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis subsp israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci U S A. 2005;102(51):18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferré J, Escriche B, Bel Y, Vanrie J. Biochemistry and genetics of insect resistance to Bacillus-thuringiensis insecticidal crystal proteins. Fems Microbiol Lett. 1995;132(1–2):1–7. doi: 10.1016/0378-1097(95)00271-6. [DOI] [Google Scholar]
- 5.Griffitts JS, Haslam SM, Yang TL, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 2005;307(5711):922–925. doi: 10.1126/science.1104444. [DOI] [PubMed] [Google Scholar]
- 6.Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66(8):1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vachon V, Laprade R, Schwartz JL. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: a critical review. J Invertebr Pathol. 2012;111(1):1–12. doi: 10.1016/j.jip.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71(2):255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Luna MT, Lanz-Mendoza H, Gill SS, Bravo A, Soberon M, Miranda-Rios J. An alpha-amylase is a novel receptor for Bacillus thuringiensis ssp israelensis Cry4Ba and Cry11Aa toxins in the malaria vector mosquito Anopheles albimanus (Diptera: Culicidae) Environ Microbiol. 2010;12(3):746–757. doi: 10.1111/j.1462-2920.2009.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tetreau G, Stalinski R, Kersusan D, Veyrenc S, David JP, Reynaud S, Despres L. Decreased Toxicity of Bacillus thuringiensis subsp. israelensis to Mosquito Larvae after Contact with Leaf Litter. Appl Environ Microbiol. 2012;78(15):5189–5195. doi: 10.1128/AEM.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetreau G, Alessi M, Veyrenc S, Périgon S, David JP, Reynaud S, Despres L. Fate of Bacillus thuringiensis subsp. israelensis in the field: evidence for spore recycling and differential persistence of toxins in leaf litter. Appl Environ Microbiol. 2012;78(23):8362–8367. doi: 10.1128/AEM.02088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paris M, Tetreau G, Laurent F, Lelu M, Despres L, David J-P. Persistence of Bacillus thuringiensis israelensis (Bti) in the environment induces resistance to multiple Bti toxins in mosquitoes. Pest Manag Sci. 2011;67:122–128. doi: 10.1002/ps.2046. [DOI] [PubMed] [Google Scholar]
- 13.Paris M, David JP, Despres L. Fitness costs of resistance to Bti toxins in the dengue vector Aedes aegypti. Ecotoxicology. 2011;20:1184–1194. doi: 10.1007/s10646-011-0663-8. [DOI] [PubMed] [Google Scholar]
- 14.Likitvivatanavong S, Chen JW, Evans AM, Bravo A, Soberon M, Gill SS. Multiple Receptors as Targets of Cry Toxins in Mosquitoes. J Agric Food Chem. 2011;59(7):2829–2838. doi: 10.1021/jf1036189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JW, Aimanova KG, Fernandez LE, Bravo A, Soberon M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp israelensis. Biochem J. 2009;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. 1988;203(2):411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- 17.Andersen SO. Amino acid sequence studies on endocuticular proteins from the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 1998;28(5–6):421–434. doi: 10.1016/S0965-1748(98)00028-9. [DOI] [PubMed] [Google Scholar]
- 18.Willis JH, Iconomidou V, Smith RF, Hamodrakas SJ. Cuticular proteins. In: Gilbert L, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science, Volume 4. Oxford: Elsevier; 2005. pp. 79–110. [Google Scholar]
- 19.Jurat-Fuentes JL, Karumbaiah L, Jakka SRK, Ning CM, Liu CX, Wu KM, Jackson J, Gould F, Blanco C, Portilla M, Perera O, Adang M. Reduced Levels of Membrane-Bound Alkaline Phosphatase Are Common to Lepidopteran Strains Resistant to Cry Toxins from Bacillus thuringiensis. PLoS One. 2011;6(3):e17606. doi: 10.1371/journal.pone.0017606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karumbaiah L, Oppert B, Jurat-Fuentes JL, Adang MJ. Analysis of midgut proteinases from Bacillus thuringiensis-susceptible and -resistant Heliothis virescens (Lepidoptera : Noctuidae) Comp Biochem Physiol B-Biochem Mol Biol. 2007;146(1):139–146. doi: 10.1016/j.cbpb.2006.10.104. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem. 2002;277(49):46849–46851. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- 22.Tetreau G, Bayyareddy K, Jones CM, Stalinski R, Riaz MA, Paris M, David JP, Adang MJ, Despres L. Larval Midgut Modifications Associated with Bti Resistance in the Yellow Fever Mosquito using Proteomic and Transcriptomic Approaches. BMC Genomics. 2012;13:248. doi: 10.1186/1471-2164-13-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tetreau G, Stalinski R, David JP, Despres L. Increase in larval gut proteolytic activities and Bti resistance in the dengue fever mosquito. Arch Insect Biochem Physiol. 2013;82(2):71–83. doi: 10.1002/arch.21076. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Cabrera L, Trujillo-Bacallao D, Borras-Hidalgo O, Wright DJ, Ayra-Pardo C. RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ Microbiol. 2010;12(11):2894–2903. doi: 10.1111/j.1462-2920.2010.02259.x. [DOI] [PubMed] [Google Scholar]
- 25.Bayyareddy K, Zhu X, Orlando R, Adang MJ. Proteome Analysis of Cry4Ba Toxin-interacting Aedes aegypti Lipid Rafts using geLC-MS/MS. J Proteome Res. 2012;11(12):5843–5855. doi: 10.1021/pr3006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rietveld A, Neutz S, Simons K, Eaton S. Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J Biol Chem. 1999;274(17):12049–12054. doi: 10.1074/jbc.274.17.12049. [DOI] [PubMed] [Google Scholar]
- 27.Avisar D, Segal M, Sneh B, Zilberstein A. Cell-cycle-dependent resistance to Bacillus thuringiensis Cry1C toxin in Sf9 cells. J Cell Sci. 2005;118(14):3163–3171. doi: 10.1242/jcs.02440. [DOI] [PubMed] [Google Scholar]
- 28.Pust S, Dyve AB, Torgersen ML, van Deurs B, Sandvig K. Interplay between Toxin Transport and Flotillin Localization. Plos One. 2010;5(1):e8844. doi: 10.1371/journal.pone.0008844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iconomidou VA, Willis JH, Hamodrakas SJ. Unique features of the structural model of ‘hard’ cuticle proteins: implications for chitin-protein interactions and cross-linking in cuticle. Insect Biochem Mol Biol. 2005;35(6):553–560. doi: 10.1016/j.ibmb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. 2010;40(3):214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Christensen BM, Li JY, Chen CC, Nappi AJ. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005;21(4):192–199. doi: 10.1016/j.pt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Hillyer JF, Christensen BM. Mosquito phenoloxidase and defensin colocalize in melanization innate immune responses. J Histochem Cytochem. 2005;53(6):689–698. doi: 10.1369/jhc.4A6564.2005. [DOI] [PubMed] [Google Scholar]
- 33.Koella JC, Boete C. A genetic correlation between age at pupation and melanization immune response of the yellow fever mosquito Aedes aegypti. Evolution. 2002;56(5):1074–1079. doi: 10.1111/j.0014-3820.2002.tb01419.x. [DOI] [PubMed] [Google Scholar]
- 34.Xie RY, Zhuang MB, Ross LS, Gomez I, Oltean DI, Bravo A, Soberon M, Gill SS. Single amino acid mutations in the cadherin receptor from Heliothis virescens affect its toxin binding ability to Cry1A toxins. J Biol Chem. 2005;280(9):8416–8425. doi: 10.1074/jbc.M408403200. [DOI] [PubMed] [Google Scholar]
- 35.Siqueira HAA, Gonzalez-Cabrera J, Ferré J, Flannagan R, Siegfried BD. Analyses of Cry1Ab binding in resistant and susceptible strains of the European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Crambidae) Appl Environ Microbiol. 2006;72(8):5318–5324. doi: 10.1128/AEM.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayed A, Nekl ER, Siqueira HAA, Wang HC, Ffrench-Constant RH, Bagley M, Siegfried BD. A novel cadherin-like gene from western corn rootworm, Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae), larval midgut tissue. Insect Mol Biol. 2007;16:591–600. doi: 10.1111/j.1365-2583.2007.00755.x. [DOI] [PubMed] [Google Scholar]
- 37.Peng DH, Xu XH, Ye WX, Yu ZN, Sun M. Helicoverpa armigera cadherin fragment enhances Cry1Ac insecticidal activity by facilitating toxin-oligomer formation. Appl Microbiol Biotechnol. 2010;85(4):1033–1040. doi: 10.1007/s00253-009-2142-1. [DOI] [PubMed] [Google Scholar]
- 38.Likitvivatanavong S, Chen JW, Bravo A, Soberon M, Gill SS. Cadherin, Alkaline Phosphatase, and Aminopeptidase N as Receptors of Cry11Ba Toxin from Bacillus thuringiensis subsp. jegathesan in Aedes aegypti. Appl Environ Microbiol. 2011;77(1):24–31. doi: 10.1128/AEM.01852-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua G, Zhang R, Abdullah MAF, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry. 2008;47(18):5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saengwiman S, Aroonkesorn A, Dedvisitsakul P, Sakdee S, Leetachewa S, Angsuthanasombat C, Pootanakit K. In vivo identification of Bacillus thuringiensis Cry4Ba toxin receptors by RNA interference knockdown of glycosylphosphatidylinositol-linked aminopeptidase N transcripts in Aedes aegypti larvae. Biochem Biophys Res Commun. 2011;407(4):708–713. doi: 10.1016/j.bbrc.2011.03.085. [DOI] [PubMed] [Google Scholar]
- 41.Flannagan RD, Yu CG, Mathis JP, Meyer TE, Shi XM, Siqueira HAA, Siegfried BD. Identification, cloning and expression of a Cry1Ab cadherin receptor from European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Crambidae) Insect Biochem Mol Biol. 2005;35(1):33–40. doi: 10.1016/j.ibmb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Fabrick JA, Tabashnik BE. Binding of Bacillus thuringiensis toxin Cry1Ac to multiple sites of cadherin in pink bollworm. Insect Biochem Mol Biol. 2007;37(2):97–106. doi: 10.1016/j.ibmb.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Fabrick J, Oppert C, Lorenzen MD, Morris K, Oppert B, Jurat-Fuentes JL. A Novel Tenebrio molitor Cadherin Is a Functional Receptor for Bacillus thuringiensis Cry3Aa Toxin. J Biol Chem. 2009;284(27):18401–18410. doi: 10.1074/jbc.M109.001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochimica Et Biophysica Acta-Biomembranes. 2004;1667(1):38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Morin S, Biggs RW, Sisterson MS, Shriver L, Ellers-Kirk C, Higginson D, Holley D, Gahan LJ, Heckel DG, Carriere Y, Dennehy TJ, Brown JK, Tabashnik BE. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc Natl Acad Sci U S A. 2003;100(9):5004–5009. doi: 10.1073/pnas.0831036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JW, Aimanova KG, Pan SQ, Gill SS. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009;39(10):688–696. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh A, Sivaprasad C. Functional interpretation of APN receptor from M.sexta using a molecular model. Bioinformation. 2009;3(8):321–325. doi: 10.6026/97320630003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 50.Tiňo P. Basic properties and information theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC Bioinformatics. 2009;10:310. doi: 10.1186/1471-2105-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paris M, Marcombe S, Coissac E, Corbel V, David JP, Despres L. Investigating the genetics of Bti resistance using mRNA tag sequencing: application on laboratory strains and natural populations of the dengue vector Aedes aegypti. Evol Appl. 2013;6(7):1012–1027. doi: 10.1111/eva.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Sequencing and mapping statistics. (XLSX 15 KB)
Additional file 2: Table S2: Blast2GO analysis of all predicted peptides annotated as ‘conserved hypothetical protein’ or ‘hypothetical protein’ in Vectorbase against the protein database Swissprot (Blastp, E-value < 10-3, annotation cut-off = 55, GO weight = 5). (XLSX 134 KB)
Additional file 3: Table S3: List of genes differently expressed between resistant and susceptible Bora-Bora strains classified according to their category. (XLSX 2 MB)
Additional file 4: Table S4: GO terms enrichment analysis. Analysis was performed on transcripts significantly differentially expressed in each LiTOX phenotype as compared to the susceptible strain. GO terms associated with each transcript were extracted from Vectorbase. GO terms showing adjusted P values <0.05 were considered significantly enriched. (XLSX 17 KB)
Additional file 5: Table S5: List of supercontigs and genes affected by differential SNPs and their effects. The total number of SNPs affecting these genes is also shown. (XLSX 315 KB)
Additional file 6: Table S6: List of genes with differential SNPs between LiTOX_N and LiTOX_S; non-synonymous changes are indicated. (XLSX 19 KB)
Additional file 7: Figure S1: RPKM correlations between cDNA library replicates. Each dot represents one transcript. Only transcripts showing more than 0.5 RPKM are shown. (XLSX 2 MB)


