Figure 3.
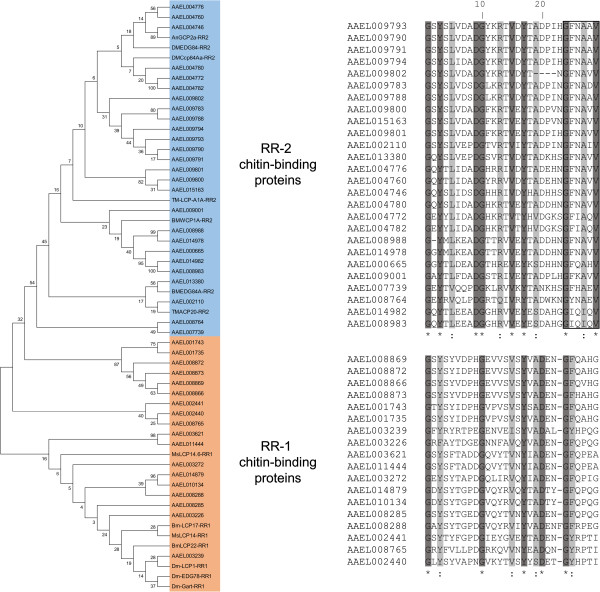
Phylogenetic analysis and sequence alignment of R&R consensus domain from the 44 differentially transcribed genes encoding chitin-binding proteins. Representatives from the two RR sub-groups from other insect species were added to support the tree (Anopheles gambiae: AnGCP2a-RR2, AAC05656; Bombyx mori: Bm-LCP17-RR1, FAA00504; BmLCP22-RR1, NP_001036828; BMEDG84A-RR2, BAA33195; BMWCP1A-RR2, BAB32475; Drosophila melanogaster: Dm-LCP1-RR1, NP_476619; Dm-EDG78-RR1, NP_524198; Dm-Gart-RR1, NP_476673; DMCcp84Aa-RR2, AAD19803; DMEDG84-RR2, P27780; Manduca sexta: MsLCP14-RR1, AAA29319; MsLCP14.6-RR1, Q94984; Tenebrio molitor: TM-LCP-A1A-RR2, P80681; TMACP20-RR2, P26967. Bootstrap values (2000 replicates) are shown on each branch of the tree. Symbols below the aligned amino acids indicate identity (*), or high conservation (:). Identical and highly conserved amino acids are highlighted in dark grey and light grey, respectively. The GFxAxV motif, diagnostic of the RR-2 sub-group, is highlighted by a box.
