Abstract
Hypoxemia is a major complication of chronic obstructive pulmonary disease (COPD) that correlates with disease prognosis. Identifying genetic variants associated with oxygenation may provide clues for deciphering the heterogeneity in prognosis among patients with COPD. However, previous genetic studies have been restricted to investigating COPD candidate genes for association with hypoxemia. To report results from the first genome-wide association study (GWAS) of resting oxygen saturation (as measured by pulse oximetry [Spo2]) in subjects with COPD, we performed a GWAS of Spo2 in two large, well characterized COPD populations: COPDGene, including both the non-Hispanic white (NHW) and African American (AA) groups, and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE). We identified several suggestive loci (P < 1 × 10−5) associated with Spo2 in COPDGene in the NHW (n = 2810) and ECLIPSE (n = 1758) groups, and two loci on chromosomes 14 and 15 in the AA group (n = 820) from COPDGene achieving a level of genome-wide significance (P < 5 × 10−8). The chromosome 14 single-nucleotide polymorphism, rs6576132, located in an intergenic region, was nominally replicated (P < 0.05) in the NHW group from COPDGene. The chromosome 15 single-nucleotide polymorphisms were rare in subjects of European ancestry, so the results could not be replicated. The chromosome 15 region contains several genes, including TICRR and KIF7, and is proximal to RHCG (Rh family C glyocoprotein gene). We have identified two loci associated with resting oxygen saturation in AA subjects with COPD, and several suggestive regions in subjects of European descent with COPD. Our study highlights the importance of investigating the genetics of complex traits in different racial groups.
Keywords: chronic obstructive pulmonary disease, hypoxemia, pulse oximetry, genome-wide association study, oxygen saturation
Clinical Relevance
Hypoxemia is a major complication of chronic obstructive pulmonary disease (COPD) that is correlated with increased risk of mortality. From our analysis of the first genome-wide association study of resting oxygen saturation in COPD, we have highlighted genes and pathways that may provide clues for deciphering heterogeneity in prognosis among patients with COPD.
Chronic obstructive pulmonary disease (COPD) is a major cause of disability, and the third leading cause of death in the United States (1). Chronic hypoxemia is a strong predictor of mortality in COPD (2, 3). However, its relationship with the severity of airflow limitation is weak. We hypothesized that identifying genetic variants associated with the severity of hypoxemia could help decipher the heterogeneity in COPD.
Blood oxygen saturation has been termed the “fifth vital sign,” and can be easily measured by pulse oximetry (Spo2) in clinical settings (4). Spo2 is an indirect measure of arterial oxygen saturation (5). Low Spo2 values are generally caused by abnormalities of pulmonary gas exchange and, in rare cases, by mutations in hemoglobin proteins (6, 7). Furthermore, genetic variants in several genes have been associated with oxygen saturation in individuals living at high altitudes (8–10). Thus, similar to many other genetic traits, there is evidence of both Mendelian and complex determinants of resting oxygen saturation. This motivated us to investigate the genetic etiology of resting oxygen saturation in subjects with COPD through a genome-wide association of resting oxygen saturation in two large cohorts of well characterized patients with COPD (COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints [ECLIPSE]).
Materials and Methods
Study Participants
Details of the COPDGene and ECLIPSE studies have been previously described (11–13). Briefly, the COPDGene Study (NCT00608764, www.copdgene.org) enrolled non-Hispanic white (NHW) and African American (AA) subjects with COPD and control subjects, aged 45–80 years, with at least 10 pack-years of lifetime smoking history in 21 U.S. centers (14). The ECLIPSE study (SCO104960, NCT00292552; www.eclipse-copd.com) recruited patients with COPD and control subjects aged 45–75 years with a smoking history of at least 10 pack-years from 46 centers across 12 countries (11). Subjects with other significant lung diseases were excluded from both studies, although COPDGene did not exclude enrollment of subjects with a history of asthma. Both COPDGene and ECLIPSE had a clinical center in Denver, Colorado at an altitude of 1.6 km above sea level. Analyses were limited to patients with COPD, defined by post-bronchodilator FEV1/forced vital capacity less than 0.7 and an FEV1 less than 80% predicted (Global Initiative for Chronic Obstructive Lung Disease grade 2 or greater) (15).
Oxygen Saturation
Spo2 was measured on a finger without nail polish after the subject had remained seated for at least 5 minutes in COPDGene, and at least 10 minutes in ECLIPSE, and recorded only if a strong pulse was apparent. The median value was recorded after observing the pulse oximetry over a 1-minute period. Subjects treated with supplemental oxygen discontinued it while being monitored with the oximeter. In the analysis for both studies, if the pulse oximeter reported a reading of less than 82%, an Spo2 value of 82% was recorded (14).
Genetic Markers
Standard quality control steps were performed on DNA samples and single-nucleotide polymorphism (SNP) data as previously described (12–14, 16). Genotyping for the COPDGene population was performed using the Illumina Human Omni 1-Quad (Illumina, San Diego, CA), and for the ECLIPSE population the Illumina HumanHap 550v3 chips were used. The COPDGene data have been deposited into dbGaP (accession number phs000179.v1.p1). For both studies, additional genotypes were imputed using the 1,000 Genomes Reference panel (17). Only SNPs with an imputation quality score of 0.8 or greater were included in the analysis. A total of 4,749,595 imputed variants in the NHW subjects with COPD from COPDGene, 4,753,821 imputed variants in ECLIPSE subjects with COPD, and 6,160,662 imputed variants in the AA subjects with COPD from COPDGene passed quality control.
Statistical Analysis
Unless otherwise specified, statistical analyses were performed in R or PLINK (18). Spo2 values were log transformed to normalize their frequency distribution. Each SNP with minor allele frequency (MAF) of 5% or greater was tested for its correlation with resting oxygenation, as measured by the log10-transformed Spo2, using an additive model adjusted for age, sex, pack-years of smoking, Denver as a study site, and principal components to summarize genetic background. Sensitivity analyses were performed by adjusting for FEV1 and/or current smoking, and also by excluding participants from Denver. Analyses were performed in the COPDGene NHW and AA groups and the ECLIPSE study separately. The initial analysis was performed in NHW cases from COPDGene, the largest group, with replication in ECLIPSE. We then investigated the COPDGene AA cases while using the other two populations as replication samples. For the loci reaching genome-wide significance (GWS) in the AA population from COPDGene, we used HaploReg2 (19), with an r2 value of 0.8 based on linkage disequilibrium information of 1,000 Genomes African Ancestry population, to assess evidence that SNPs were located in regulatory regions. Meta-analyses were performed between NHW cases from COPDGene and ECLIPSE, in addition to all three populations, including the AA subjects from COPDGene. Gene-based analyses were performed using VEGAS (20), software that generates a gene-based result based on single SNP P values from the genome-wide association study (GWAS) and linkage disequilibrium patterns (20). VEGAS assigns SNPs to genes based on physical position (± 50 kb) of known genes in the University of California, Santa Cruz (Santa Cruz, CA) genome browser. Regional association plots were generated using LocusZoom (21).
Results
Characterization of Participants
Table 1 presents the main clinical characteristics of the participants represented in this investigation. In ECLIPSE, subjects with COPD consisted of a higher proportion of males (67.2%) than COPDGene NHW and AA cases (55.7 and 55.1%, respectively). The AA case population was, on average, slightly younger and also had a lower mean exposure to smoking, as measured by pack-years, than the NHW and ECLIPSE cases. The ECLIPSE subjects were, on average, slightly less overweight (mean body mass index = 26.7 kg/m2), but had lower lung function measurements than subjects with COPD from the COPDGene study. ECLIPSE also had fewer participants recruited from Denver than either COPDGene case populations. The percentage of subjects with COPD with severe hypoxemia, defined as Spo2 of 88% or less, was higher in the NHW (7%) than both the AA and ECLIPSE cases (4%). However, the median and range of Spo2 in each population were comparable with the medians for all three study populations, centered at approximately 95%. We also examined the subset of subjects within each study recruited from Denver, the site with the highest altitude (see Table E1 in the online supplement). Subjects recruited from Denver were slightly less overweight and had lower lung function than the respective total case populations. Furthermore, the median percent Spo2 was lower and there were more cases with severe hypoxemia recruited from Denver compared with the total study populations.
Table 1.
Characteristics of Subjects with Chronic Obstructive Pulmonary Disease Included in the Analysis
| COPDGene |
|||
|---|---|---|---|
| Characteristic | Non-Hispanic White | African American | ECLIPSE |
| n | 2,810 | 820 | 1,758 |
| Males, % | 55.7 | 55.1 | 67.2 |
| Age, yr | 64.7 (8.2) | 59 (8.2) | 63.6 (7.1) |
| BMI, kg/m2 | 28.1 (6.1) | 28 (6.8) | 26.7 (5.6) |
| Smoking exposure, pack-years | 56.3 (28) | 42.4 (23) | 50.3 (27) |
| Enrollment in Denver, % | 22.7 | 4.3 | 1.2 |
| FEV1 % predicted | 49.6 (18) | 52.2 (17.8) | 43.5 (14.9) |
| FEV1/FVC, % | 0.49 (0.13) | 0.53 (0.12) | 0.44 (0.11) |
| Spo2, %* | 95 (17) | 97 (17) | 95 (18) |
| Severe hypoxemia, %† | 7 | 4 | 4 |
Definition of abbreviations: BMI, body mass index; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study; FVC, forced vital capacity; Spo2, oxygen saturation measured using pulse oximetry.
Values presented are mean and SD unless otherwise noted.
Median (range).
Spo2 ≤ 88%.
Distribution of Spo2 in Subjects with COPD
The distribution of Spo2 in subjects with COPD in COPDGene and ECLIPSE was not normally distributed (see Figure E1). As a result, a log transformation was used to normalize the trait. Figure E1 displays the distribution of residuals of the log-transformed trait after adjusting for sex, lifetime smoking intensity, age, Denver site, and population substructure for each COPD case population.
GWAS of Spo2 in Subjects with COPD with European Ancestry
In the GWAS of the NHW population from COPDGene, no SNP reached GWS in tests of association with log(Spo2), nor did any SNP reach GWS in the meta-analysis between the COPDGene and ECLIPSE (data not shown). In Table 2, the top 10 SNPs associated with log(Spo2) sorted by chromosome and physical position are listed for the COPDGene NHW and ECLIPSE analyses. Several SNPs on chromosome 1, approximately 71 kb 5′ of the gene, RUNX3 (runt-related transcription factor 3), were associated (P = 1.2 × 10−6) with log(Spo2) in the NHW group in COPDGene. Several SNPs near NRXN1 (neurexin-1-α) on chromosome 2 were associated with log(Spo2) (P < 7.1 × 10−7). SNP rs6893408 on chromosome 5 located within the FGF1 (fibroblast growth factor 1) gene was associated with log(Spo2) in COPDGene (Beta = −0.0026 log[Spo2] units; P = 4.8 × 10−7), which corresponded to a mean difference in Spo2 of −1.0% Spo2 for each minor allele. Three SNPs, with TRIQK (triple QxxK/R motif containing) as the closest gene, were moderately associated with resting oxygenation in the NHW population. Several SNPs in the CBR4 (carnbonyl reductase family member 4) gene region on chromosome 4 and single SNPs in the genes, OPCML (Opioid Binding Protein/Cell Adhesion) and BRSK2 (BR serine/threonine kinase 2), were associated with log(Spo2) in the ECLIPSE cases; however, these results did not reach GWS. The top results in COPDGene NHW and ECLIPSE subjects were examined in the other populations (Tables E2 and E3). As a secondary analysis, we also tested variants in the two largest populations, COPDGene NHW and ECLIPSE, with MAF greater than 1%. No variant reached a level of GWS in the largest population: the NHW from COPDGene. However, one variant with a MAF of 2.3%, rs7868621 on chromosome 9, reached GWS (Beta = −0.0080; SE = 0.0014; P = 4.3 × 10−8) in ECLIPSE. This variant did not replicate in either COPDGene population (PNHW = 0.80 and PAA = 0.21, respectively).
Table 2.
Top 10 Single-Nucleotide Polymorphisms Associated with log10(SpO2) in Non-Hispanic White Subjects with Chronic Obstructive Pulmonary Disease from the COPDGene and ECLIPSE Studies
| Excluding Participants from Denver |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | MAF | ||||||||||||||||
| CHR | SNP | Position | (%) | BETA | 95% CI | P Value | (%) | BETA | 95% CI | P Value | Closest Gene | ||||||
| COPDGene non-Hispanic white subjects with COPD (n = 2,810) |
|||||||||||||||||
| 1 | rs10903128 | 25,363,338 | 27.7 | −0.0020 | −0.0028 to −0.0012 | 1.2E−06 | 26.9 | −0.0010 | −0.0018 to −0.0002 | 0.013 | RUNX3 (∼72 kB) | ||||||
| 1 | rs452044 | 25,363,656 | 27.9 | −0.0020 | −0.0028 to −0.0012 | 1.2E−06 | 27.8 | −0.0010 | −0.0017 to −0.0002 | 0.015 | RUNX3 (∼72 kB) | ||||||
| 1 | rs453532 | 25,363,707 | 27.7 | −0.0020 | −0.0028 to −0.0012 | 1.2E−06 | 27.0 | −0.0010 | −0.0018 to −0.0002 | 0.013 | RUNX3 (∼72 kB) | ||||||
| 2 | rs10190452 | 52,230,237 | 45.8 | 0.0019 | 0.0011 to 0.0027 | 4.3E−07 | 45.7 | 0.0013 | 0.0007 to 0.0019 | 1.2E−04 | NRXN1 (∼97 kB) | ||||||
| 2 | rs6736403 | 52,233,463 | 45.6 | 0.0019 | 0.0011 to 0.0026 | 4.1E−07 | 45.7 | 0.0013 | 0.0006 to 0.0020 | 1.5E−04 | NRXN1 (∼97 kB) | ||||||
| 2 | rs7566343 | 52,240,977 | 44.9 | 0.0018 | 0.0010 to 0.0026 | 7.1E−07 | 44.8 | 0.0014 | 0.0008 to 0.0020 | 8.8E−05 | NRXN1 (∼97 kB) | ||||||
| 5 | rs6893408 | 142,022,777 | 14.3 | −0.0026 | −0.0036 to −0.0016 | 4.8E−07 | 14.1 | −0.0012 | −0.0022 to −0.0003 | 0.014 | FGF1 | ||||||
| 8 | rs536161 | 93,513,027 | 27.8 | −0.0019 | −0.0027 to −0.0011 | 2.5E−06 | 28.4 | −0.0010 | −0.0018 to −0.0002 | 0.013 | TRIQK (∼380 kB) | ||||||
| 8 | rs511954 | 93,514,434 | 27.9 | −0.0019 | −0.0027 to −0.0011 | 2.4E−06 | 28.4 | −0.0010 | −0.0018 to −0.0002 | 0.013 | TRIQK (∼380 kB) | ||||||
| 8 | rs482619 | 93,514,906 | 28.2 | −0.0019 | −0.0028 to −0.0011 | 2.4E−06 | 28.1 | −0.0010 | −0.0017 to −0.0002 | 0.013 | TRIQK (∼380 kB) | ||||||
| ECLIPSE subjects with COPD (n = 1,758) |
|||||||||||||||||
| 4 | rs115325116 | 169,933,895 | 6.4 | −0.0047 | −0.0065 to −0.0029 | 2.3E−07 | 6.4 | −0.0047 | −0.006 to −0.003 | 2.6E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs180977786 | 169,933,903 | 6.6 | −0.0047 | −0.0065 to −0.0029 | 2.4E−07 | 6.6 | −0.0047 | −0.006 to −0.003 | 2.7E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs17615397 | 169,937,521 | 5.7 | −0.0050 | −0.0070 to −0.0030 | 3.8E−07 | 5.7 | −0.0050 | −0.007 to −0.003 | 4.2E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs74581741 | 169,940,336 | 5.7 | −0.0050 | −0.0070 to −0.0030 | 3.8E−07 | 5.7 | −0.0050 | −0.007 to −0.003 | 4.2E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs17615362 | 169,934,087 | 6.4 | −0.0046 | −0.0064 to −0.0028 | 4.0E−07 | 6.3 | −0.0046 | −0.006 to −0.003 | 4.4E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs17543620 | 169,934,725 | 6.4 | −0.0046 | −0.0064 to −0.0028 | 4.0E−07 | 6.3 | −0.0046 | −0.006 to −0.003 | 4.5E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs148456540 | 169,937,248 | 5.4 | −0.0052 | −0.0072 to −0.0032 | 4.4E−07 | 5.4 | −0.0051 | −0.007 to −0.003 | 5.3E−07 | CBR4 (∼2 kB) | ||||||
| 4 | rs77404015 | 169,934,424 | 6.5 | −0.0044 | −0.0062 to −0.0026 | 1.0E−06 | 6.5 | −0.0044 | −0.006 to −0.003 | 1.1E−06 | CBR4 (∼2 kB) | ||||||
| 11 | rs4379857 | 133,183,604 | 5.5 | −0.0045 | −0.0063 to −0.0026 | 2.2E−06 | 5.5 | −0.0045 | −0.006 to −0.003 | 2.0E−06 | OPCML | ||||||
| 11 | rs138091420 | 1,399,692 | 8.3 | −0.0038 | −0.0054 to −0.0022 | 2.3E−06 | 8.3 | −0.0036 | −0.005 to −0.002 | 6.0E−06 | BRSK2 (11 kB) | ||||||
Definition of abbreviations: CHR, chromosome; CI, confidence interval; COPD, chronic obstructive pulmonary disease; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study; MAF, minor allele frequency; SNP, single-nucleotide polymorphism; SpO2, oxygen saturation measured using pulse oximetry.
The term “Position” indicates the Human Genome Build 19 physical position in base pairs, and “Closest gene” is the closest gene to the SNP, where SNP distance to gene is denoted in parentheses when SNP is not within gene.
We also examined the top results adjusting for FEV1, current smoking, and both together (Table E4). The top results did not change substantially when FEV1 and/or current smoking were considered. Furthermore, we found that the top associations in ECLIPSE were robust to excluding participants from Denver (Table 2). However, after excluding cases from Denver (22.7% of the total NHW case population), the top results in the NHW population became less significant.
GWAS of Spo2 in AA Subjects with COPD
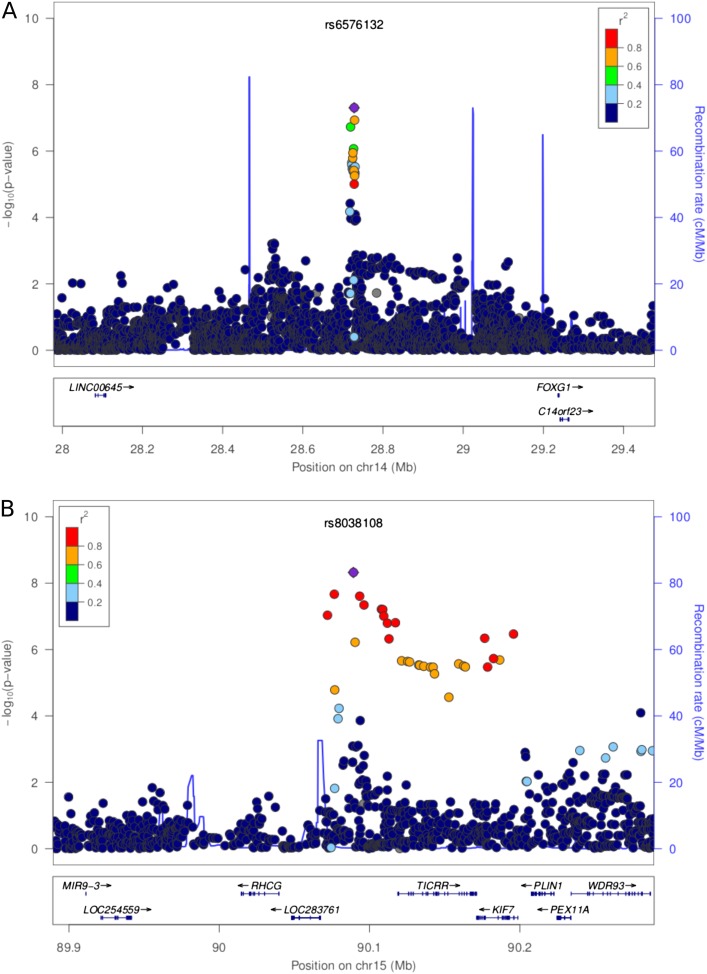
There were two SNPs with a MAF greater than 5% that were significantly associated with log(Spo2) among the COPDGene AA cases (Table 3). Further examination of variants in these regions with MAF less than 5% identified three more variants on chromosome 15 significantly associated with Spo2 (MAF, ∼4%). The chromosome 14 SNP, rs6576132 (Beta = 0.0039; P = 5.0 × 10−8), replicated nominally among the COPDGene NHW case population (Beta = 0.0009; P = 0.011). This Beta corresponded to a mean difference of 1.0% Spo2 for each minor allele. Figure E3A illustrates Spo2 stratified by rs6576132 genotype. The minor allele for this SNP is common, with an MAF of 43.4%. Approximately 145 AA subjects with COPD carried the CC genotype, with a median Spo2 of 97.7%. This is in comparison to roughly 250 AA subjects with COPD who carried the AA genotype, with a median Spo2 of 95.5%. However, there were no genes annotated to this region as is depicted in the regional association plot (Figure 1A). The five SNPs associated with log(Spo2) on chromosome 15 had MAFs that ranged from 4.2 to 5.5% in the AA population, but these markers were rare in European populations (MAF = 0.02% in COPDGene NHW cases). For this reason, we were unable to replicate this region in the NHW or ECLIPSE populations (Table E5). We assessed whether the top SNPs in Table 3 were located in regulatory regions using HaploReg2. Several of these SNPs, chr14:rs6576132, chr15: rs116033091, and chr15: rs8038108, are predicted to alter regulatory motifs. No additional loci reached GWS in the meta-analysis of all three populations (data not shown).
Table 3.
SNPs Associated at a Level of Genome-Wide Significance* with log10(SpO2) in the African American Population from COPDGene
| COPDGene African American Subjects with COPD |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 820) |
Excluding Participants from Denver |
||||||||||
| |
|
|
MAF |
|
|
|
MAF |
|
|
|
|
| CHR | SNP | Position | (%) | BETA | 95% CI | P Value | (%) | BETA | 95% CI | P Value | Closest Gene |
| 14 | rs6576132 | 28,728,111 | 43.4 | 0.004 | 0.003 to 0.005 | 4.9E−08 | 43.6 | 0.003 | 0.002 to 0.005 | 1.3E−06 | FOXG1 (∼ 508 kB) |
| 15 | rs8038108 | 90,089,276 | 4.8 | −0.009 | −0.013 to −0.006 | 4.8E−09 | 4.6 | −0.009 | −0.012 to −0.006 | 1.7E−08 | LINC00928 (∼ 22 kB) |
| 15 | rs116033091 | 90,076,553 | 4.6 | −0.009 | −0.012 to −0.006 | 2.2E−08 | 4.4 | −0.008 | −0.011 to −0.005 | 2.1E−07 | LINC00928 (∼ 9 kB) |
| 15 | rs8025537 | 90,093,403 | 4.2 | −0.009 | −0.012 to −0.006 | 2.5E−08 | 4.5 | −0.008 | −0.011 to −0.005 | 6.0E−08 | TICRR (∼ 25 kB) |
| 15 | rs147566087 | 90,096,263 | 5.5 | −0.008 | −0.011 to −0.005 | 4.6E−08 | 5.3 | −0.008 | −0.011 to −0.005 | 7.8E−08 | TICRR (∼ 25 kB) |
Definition of abbreviations: CHR, chromosome; CI, confidence interval; COPD, chronic obstructive pulmonary disease; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study; MAF, minor allele frequency; SNP, single-nucleotide polymorphism; SpO2, oxygen saturation measured using pulse oximetry.
The term “Position” indicates the Human Genome Build 19 physical position in base pairs, and “Closest gene” is the closest gene to the SNP, where SNP distance to gene is denoted in parentheses when SNP is not within gene.
P ≤ 5.0 × 10−8.
Figure 1.
Regional association plots around single-nucleotide polymorphisms reaching genome-wide significance for association with log(SpO2) in African American subjects from COPDGene. (A) Chromosome 14; (B) chromosome 15. SpO2, oxygen saturation measured using pulse oximetry.
Figures E3A and E3B show Spo2 stratified by genotype for the single chromosome 14 SNP and two chromosome 15 SNPs exhibiting GWS. The KIF7 (kinesin family member 7) gene is annotated to the chromosome 15 region (Figure 1B). This region is also in close proximity to the RHCG (Rh family, C glycoprotein) gene; however, the genetic region associated with log(Spo2) in AA subjects was separated from RHCG by a recombination hot spot. Similar to the analysis in the two populations of European descent, adjusting for the effect of FEV1 and/or current smoking in the AA cases had no major effect on the top results (Table E6). However, when subjects recruited from Denver (4.3% of the total AA case population) were excluded from the analysis, the association on chromosome 14 no longer reached GWS (Beta = 0.003; P = 1.3 × 10−6; Table 3). However, the association with rs8038108 on chromosome 15 remained of GWS (Beta = −0.009; P = 1.7 × 10−8).
Gene-Based Associations with Spo2 in COPD
We next performed a gene-based analysis for 17,640 genes, as opposed to a single SNP–based analysis, in each population using VEGAS. None of the gene-based results withstood Bonferonni correction (P < 2.8 × 10−6). The top results from each population are listed in Table E7. For comparison, we also examined the results excluding participants from Denver. In the COPDGene NHW population, the top gene was BIN1 on chromosome 2 (Ptop20% = 9.0 × 10−6). The next highest-ranking gene was TGFBR3, which our group has previously shown to be associated with emphysema (22). The top gene associated with log(Spo2) in ECLIPSE was CLEC12A (Ptop20% = 3.0 × 10−5). The top gene in the AA population was HSD17B3 (Ptop20% = 1.1 × 10−5). We also looked specifically for associations with hemoglobin genes in the hemoglobin α and β gene regions on chromosomes 11 and 16 in each population (Table E8). Interestingly, none of these genes reached nominal significance (P < 0.05) in the NHW case population. However, HBA2 (Ptop20% = 0.039) and HBM (Ptop20% = 0.044) were nominally significant in the AA COPD case population.
Discussion
This study has identified two genetic regions that are significantly associated with resting oxygen saturation among AA subjects with COPD. We were able to replicate nominally (P < 0.05) the association on chromosome 14 in the NHW subjects from COPDGene. The SNPs we found associated on chromosome 15 with Spo2 in AA subjects with COPD were rare in populations of European descent. However, this region encompassed several candidate genes for further investigation, including TICRR, KIF7, and RHCG. Although we identified several suggestive associations using the two populations of European descent, NHW subjects from COPDGene and ECLIPSE, our investigation underscores the importance of investigating the genetics of complex traits in additional racial groups, such as AA subjects.
Previous Studies
Previous genetic studies of oxygen saturation in COPD focused on genes known to influence COPD susceptibility rather than oxygen saturation per se (23, 24). On the other hand, there have been few investigations into the genetics of oxygen saturation in other respiratory diseases, and these have mainly been restricted to candidate gene studies. Schroder and colleagues (25) found evidence for association of serotonin transporter polymorphism with decreased arterial oxygen saturation, and also with increasing apnea–hypopnea index in older adults. de Lima Marson and colleagues (26) searched for cystic fibrosis modifier genes by assessing genetic interactions with cystic fibrosis transmembrane conductance regulator gene mutations, but did not find a significant association with oxygen saturation itself. The role of environmental factors, such as altitude, oxygen use, exercise capacity, and quality of life, has been investigated in relation to hypoxemia in patients with COPD (27). Here, we report results from the first GWAS of resting oxygen saturation in patients with COPD.
Interpretation of Findings
The median and range of Spo2 in the three COPD case populations were similar at roughly 95%. The NHW population had a higher percentage of subjects with COPD with severe hypoxemia at 7% in comparison with 4% in the AA and ECLIPSE groups. This difference is slightly lower than the prevalence Kim and colleagues (27) reported for the prevalence of severe hypoxemia of 7.7% (n = 1,061) in the first 2,500 subjects with COPD enrolled in COPDGene. In the current report, the NHW subjects with COPD from COPDGene had the highest percentage of subjects recruited in Denver. Furthermore, the NHW cases also had more pack-years of smoking on average than the other two case populations. Smoking history is known to influence Spo2 levels even in the general population (28). For this reason, we included these variables as covariates in the GWAS, and also assessed the effects of current smoking on the results.
Our analysis of Spo2 in two COPD case populations of European descent, the NHW subjects with COPD from COPDGene and ECLIPSE, revealed suggestive associations with variants near several genes, including BRSK2, CBR4, RUNX3, and FGF1. BRSK2 is a brain-specific kinase (29). CBR4, a carbonyl reductase gene, is thought to play a role in biosynthesis of fatty acids in the mitochondria (30). RUNX3 is a transcription factor essential for CD8 T cell development (31). A related gene, RUNX1, which also contains a Runt domain, is essential for hematopoiesis, and chromosomal rearrangements in this gene are commonly found in patients with leukemia (32, 33). In our study, we observed several variants with suggestive association near RUNX3 in the NHW COPD case population in COPDGene. The variants are approximately 71 kB from RUNX3, and there is evidence of transcriptional regulation in lymphoblastoid cells (19). Furthermore, a SNP in FGF1 demonstrated suggestive association with Spo2 in our study. FGF1 is a member of the fibroblast growth factor family, which is involved in a broad range of processes. FGF1, in particular, influences endothelial cell migration and proliferation, in addition to playing a role in angiogeneisis and wound healing (34).
Most interestingly, our analysis revealed two regions associated with resting oxygen saturation in AA subjects with COPD. The region on chromosome 14 also replicated in the NHW population from COPDGene. This region is intergenic and bounded by genes, FOXG1 (forkhead box G1) and LINC00645 (noncoding RNA645), by recombination peaks (Figure 1A). The second region on chromosome 15 tags the genes, TICRR and KIF7, and is also proximal to RHCG, but is separated by a recombination peak (Figure 1B). RHCG is an intriguing candidate gene for blood oxygen saturation, as it codes for an Rh protein, which is important for maintaining the structure of the red blood cell membrane (35). TICRR codes for the treslin protein involved with the initiation of DNA replication (36). KIF7 is an essential component of the sonic hedgehog signaling pathway, and also for the development of the diaphragm (37). Furthermore, RUNX3, for which we showed suggestive associations with resting blood oxygen in the NHW subjects with COPD, is involved with Indian Hedgehog signaling in epithelial cells in the stomach and intestine (38). The HHIP (hedgehog interacting protein) gene is associated with lung function and COPD (13, 16, 39–41); however, its contribution to the pathogenesis of COPD remains to be elucidated (38, 41). The robustness of our top findings after adjusting for FEV1 suggests that we have identified true genetic associations with resting oxygen saturation as opposed to associations with COPD severity.
We also performed a gene-based analysis in each COPD case population testing for association with resting oxygen saturation. Although no gene withstood correction for multiple testing, the gene, TGFBR3, which has been previously associated with emphysema, was among the top genes associated in the NHW subjects with COPD from COPDGene (22). We saw no evidence of association with common variants in hemoglobin genes on chromosomes 11 and 16.
Potential Limitations
Our study has several limitations. Our study lacks an AA replication population for the findings achieving GWS in the AA subjects from COPDGene. It is well established that the pattern of linkage disequilibrium between individuals of European and African descent differs (42, 43). In our report, the variants that we have identified as associated with Spo2 in the AA population on chromosome 15 were extremely rare in European populations. However, the variants in the chromosome 14 region are common in both the AA and NHW populations. For these more common variants, we were able to nominally replicate (P < 0.05) statistical associations in the larger NHW population. With complex traits, it is possible that different variants are associated in populations with different racial/ethnic populations.
Furthermore, the top result on chromosome 14 in the COPDGene AA cases became less significant when participants from Denver were excluded, but the top results on chromosome 15 were robust to excluding subjects from Denver. This indicates that the chromosome 14 finding is confounded by living at high altitude, whereas the chromosome 15 finding is not. Although genetic variants and genes have been associated adaptation to hypoxia in high-altitude populations (44–49), our results did not show overlap with these findings, which is not surprising given the differences in phenotype. Additional studies of AA subjects with COPD living at high altitude would be required to further explore the relationship between the chromosome 14 variant and resting oxygen levels.
Conclusions
This study identifies two novel regions associated with resting oxygen saturation on chromosomes 14 and 15 among AA subjects with COPD. Several genes involved in the hedgehog and transforming growth factor β pathways support the relevance of these pathways in COPD. Additional studies will be required to determine the functional variants and mechanisms by which the chromosome 14 and 15 loci could influence resting oxygen saturation in COPD. Our report underscores the importance of investigating multiple racial populations in genetic studies of complex disease.
Acknowledgments
Acknowledgments
The authors thank the participants and field investigators in both the ECLIPSE study and the COPDGene study for their willingness to contribute to medical research.
COPDGene Investigators—Core Units
Administrative Core: James Crapo, M.D. (principal investigator [PI]), Edwin Silverman, M.D., Ph.D. (PI), Barry Make, M.D., Elizabeth Regan, M.D., Rochelle Lantz, Lori Stepp, Sandra Melanson.
Genetic Analysis Core: Terri Beaty, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Peter Castaldi, M.D., M.Sc., Merry-Lynn McDonald, Ph.D., Jin Zhou, Ph.D., Manuel Mattheisen, M.D., Emily Wan, M.D., Megan Hardin, M.D., Jacqueline Hetmanski, M.S., Margaret Parker, M.S., Tanda Murray, M.S., Marilyn Foreman, M.D., M.S., Sharon M. Lutz, M.P.H., Ph.D., Elizabeth Regan, M.D., Ph.D., Edwin K. Silverman, M.D., Ph.D.
Imaging Core: David Lynch, MB, Joyce Schroeder, M.D., John Newell, Jr., M.D., John Reilly, M.D., Harvey Coxson, Ph.D., Philip Judy, Ph.D., Eric Hoffman, Ph.D., George Washko, M.D., Raul San Jose Estepar, Ph.D., James Ross, M.Sc., Mustafa Al Qaisi, M.D., Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, Douglas Stinson, Christian W. Cox, M.D., Deanna Cusick, Jennifer G. Dy, Ph.D., Shoshana Ginsburg, M.S., Arkadiusz Sitek, Ph.D., Edwin van Beek, M.D., Ph.D., M.Ed.
Pulmonary Function Testing Quality Assurance Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, Ph.D.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, Ph.D., Stacey Meyerer, Shivam Chandan, Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D., Andre Williams, Ph.D., Carla Wilson, M.S., Anna Forssen, M.S., Amber Powell, Joe Piccoli
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, M.P.H., Ph.D., Marci Sontag, Ph.D., Jennifer Black-Shinn, M.P.H., Gregory Kinney, M.P.H., Ph.D.c, Sharon Lutz, M.P.H., Ph.D.
COPDGene Investigators: Clinical Centers
Ann Arbor Veterans Affairs, Ann Arbor, MI: Jeffrey Curtis, M.D., Ella Kazerooni, M.D.
Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S., Philip Alapat, M.D., Venkata Bandi, M.D., Kalpalatha Guntupalli, M.D., Elizabeth Guy, M.D., Antara Mallampalli, M.D., Charles Trinh, M.D., Mustafa Atik, M.D., Hasan Al-Azzawi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., Anne Marie Marciel, M.D.
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H., Craig Hersh, M.D., M.P.H., George Washko, M.D., Francine Jacobson, M.D., M.P.H., Hiroto Hatabu, M.D., Ph.D., Peter Clarke, M.D., Ritu Gill, M.D., Andetta Hunsaker, M.D., Beatrice Trotman-Dickenson, M.B.B.S., Rachna Madan, M.D.
Columbia University, New York, NY: R. Graham Barr, M.D., Dr.PH., Byron Thomashow, M.D., John Austin, M.D., Belinda D’Souza, M.D.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D., Lacey Washington, M.D., H. Page McAdams, M.D.
Reliant Medical Group, Worcester, MA: Richard Rosiello, M.D., Timothy Bresnahan, M.D., Joseph Bradley, M.D., Sharon Kuong, M.D., Steven Meller, M.D., Suzanne Roland, M.D.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H., Joseph Tashjian, M.D.
Johns Hopkins University, Baltimore, MD: Robert Wise, M.D., Nadia Hansel, M.D., M.P.H., Robert Brown, M.D., Gregory Diette, M.D., Karen Horton, M.D.
Los Angeles Biomedical Research Institute at Harbor–University of California Los Angeles Medical Center, Torrance, CA: Richard Casaburi, M.D., Ph.D., Janos Porszasz, M.D., Ph.D., Hans Fischer, M.D., Matt Budoff, M.D.
Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX: Amir Sharafkhaneh, M.D., Charles Trinh, M.D., Hirani Kamal, M.D., Roham Darvishi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., Anne Marie Marciel, M.D.
Minneapolis Veterans Affairs, Minneapolis, MN: Dennis Niewoehner, M.D., Quentin Anderson, M.D., Kathryn Rice, M.D., Audrey Caine, M.D.
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S., Gloria Westney, M.D., M.S., Eugene Berkowitz, M.D., Ph.D.
National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D., David Lynch, M.B., Joyce Schroeder, M.D., Valerie Hale, M.D., John Armstrong, II, M.D., Debra Dyer, M.D., Jonathan Chung, M.D., Christian Cox, M.D.
Temple University, Philadelphia, PA: Gerard Criner, M.D., Victor Kim, M.D., Nathaniel Marchetti, D.O., Aditi Satti, M.D., A. James Mamary, M.D., Robert Steiner, M.D., Chandra Dass, M.D., Libby Cone, M.D.
University of Alabama, Birmingham, AL: William Bailey, M.D., Mark Dransfield, M.D., Michael Wells, M.D., Surya Bhatt, M.D., Hrudaya Nath, M.D., Satinder Singh, M.D.
University of California, San Diego, CA: Joe Ramsdell, M.D., Paul Friedman, M.D.
University of Iowa, Iowa City, IA: Alejandro Cornellas, M.D., John Newell, Jr., M.D., Edwin J. R. van Beek, M.D., Ph.D.
University of Michigan, Ann Arbor, MI: Fernando Martinez, M.D., Mei Lan Han, M.D., Ella Kazerooni, M.D.
University of Minnesota, Minneapolis, MN: Christine Wendt, M.D., Tadashi Allen, M.D.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D., Joel Weissfeld, M.D., M.P.H., Carl Fuhrman, M.D., Jessica Bon, M.D., Danielle Hooper, M.D.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D., Sandra Adams, M.D., Carlos Orozco, M.D., Mario Ruiz, M.D., Amy Mumbower, M.D., Ariel Kruger, M.D., Carlos Restrepo, M.D., Michael Lane, M.D.
Principal Investigators and Centers Participating in the ECLIPSE Study Include:
Bulgaria: Y. Ivanov (Pleven); K. Kostov (Sofia). Canada: J. Bourbeau (Montreal, PQ); M. Fitzgerald (Vancouver, BC); P. Hernandez (Halifax, NS); K. Killian (Hamilton, ON); R. Levy (Vancouver, BC); F. Maltais (Montreal, PQ); D. O’Donnell (Kingston, ON). Czech Republic: J. Krepelka (Prague). Denmark: J. Vestbo (Hvidovre). The Netherlands: E. Wouters (Horn–Maastricht). New Zealand: D. Quinn (Wellington). Norway: P. Bakke (Bergen). Slovenia: M. Kosnik (Golnik). Spain: A. Agusti, J. Sauleda (P. de Mallorca). Ukraine: Y. Feschenko, V. Gavrisyuk, L. Yashina (Kiev); N. Monogarova (Donetsk). United Kingdom: P. Calverley (Liverpool); D. Lomas (Cambridge); W. MacNee (Edinburgh); D. Singh (Manchester); J. Wedzicha (London). United States: A. Anzueto (San Antonio, TX); S. Braman (Providence, RI); R. Casaburi (Torrance, CA); B. Celli (Boston, MA); G. Giessel (Richmond, VA); M. Gotfried (Phoenix, AZ); G. Greenwald (Rancho Mirage, CA); N. Hanania (Houston, TX); D. Mahler (Lebanon, NH); B. Make (Denver, CO); S. Rennard (Omaha, NE); C. Rochester (New Haven, CT); P. Scanlon (Rochester, MN); D. Schuller (Omaha, NE); F. Sciurba (Pittsburgh, PA); A. Sharafkhaneh (Houston, TX); T. Siler (St. Charles, MO); E. Silverman (Boston, MA); A. Wanner (Miami, FL); R. Wise (Baltimore, MD); R. ZuWallack (Hartford, CT).
Steering Committee:
H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
Scientific Committee:
A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (The Netherlands), J. Yates (GlaxoSmithKline, USA).
Footnotes
This work was supported by National Institutes of Health grants R01 HL089856 (E.K.S.), R01HL089897 (J.D.C.), R01 HL094635 (C.P.H.), R01 NR013377 (C.P.H.), and P01 HL105339 (E.K.S.). The COPDGene study is also supported by the COPD Foundation through contributions made to an industry advisory board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, and Sunovion. The Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study was funded by GlaxoSmithKline.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0135OC on May 13, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Murphy SL, Xu J, Kochanek KD.Deaths: final data for 2010. National vital statistics reports from the Centers for Disease Control and Prevention, National Center for Health Statistics Natl Vital Stat Rep 201361(4)1–118.. Available from: http://www.cdc.gov/nchs/data/nvsr/nvsr61/nvsr61_04.pdf [PubMed] [Google Scholar]
- 2.Nizet TA, van den Elshout FJ, Heijdra YF, van de Ven MJ, Mulder PG, Folgering HT. Survival of chronic hypercapnic COPD patients is predicted by smoking habits, comorbidity, and hypoxemia. Chest. 2005;127:1904–1910. doi: 10.1378/chest.127.6.1904. [DOI] [PubMed] [Google Scholar]
- 3.Coleta KD, Silveira LV, Lima DF, Rampinelli EA, Godoy I, Godoy I. Predictors of first-year survival in patients with advanced COPD treated using long-term oxygen therapy. Respir Med. 2008;102:512–518. doi: 10.1016/j.rmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Neff TA. Routine oximetry: a fifth vital sign? Chest. 1988;94:227. doi: 10.1378/chest.94.2.227a. [DOI] [PubMed] [Google Scholar]
- 5.Chan ED, Chan MM, Chan MM. Pulse oximetry: understanding its basic principles facilitates appreciation of its limitations. Respir Med. 2013;107:789–799. doi: 10.1016/j.rmed.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Verhovsek M, Henderson MP, Cox G, Luo HY, Steinberg MH, Chui DH. Unexpectedly low pulse oximetry measurements associated with variant hemoglobins: a systematic review. Am J Hematol. 2010;85:882–885. doi: 10.1002/ajh.21810. [DOI] [PubMed] [Google Scholar]
- 7.Weil JV. Variation in human ventilatory control—genetic influence on the hypoxic ventilatory response. Respir Physiol Neurobiol. 2003;135:239–246. doi: 10.1016/s1569-9048(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 8.Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, Zhu WZ, Scott CR, Chen SH. VEGFA SNPs and transcriptional factor binding sites associated with high altitude sickness in Han and Tibetan Chinese at the Qinghai–Tibetan Plateau. J Physiol Sci. 2013;63:183–193. doi: 10.1007/s12576-013-0257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra A, Mohammad G, Thinlas T, Pasha MA. EGLN1 variants influence expression and SaO2 levels to associate with high-altitude pulmonary oedema and adaptation. Clin Sci (Lond) 2013;124:479–489. doi: 10.1042/CS20120371. [DOI] [PubMed] [Google Scholar]
- 10.Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, Zhu WZ, Scott CR, Chen SH. AKT3, ANGPTL4, eNOS3, and VEGFA associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Int J Hematol. 2012;96:200–213. doi: 10.1007/s12185-012-1117-7. [DOI] [PubMed] [Google Scholar]
- 11.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, et al. ECLIPSE investigators. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 12.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 16.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, et al. ICGN Investigators. Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. Am J Respir Cell Mol Biol. 2009;41:324–331. doi: 10.1165/rcmb.2008-0427OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castaldi PJ, Hersh CP, Reilly JJ, Silverman EK. Genetic associations with hypoxemia and pulmonary arterial pressure in COPD. Chest. 2009;135:737–744. doi: 10.1378/chest.08-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Córdoba-Lanús E, de-Torres JP, López-Aguilar C, Rodríguez-Pérez MC, Maca-Meyer N, Montejo-de-Garcini A, Aguirre-Jaime A, Pérez-Méndez L, Casanova C. Association of IL-6 gene polymorphisms and COPD in a Spanish population. Respir Med. 2008;102:1805–1811. doi: 10.1016/j.rmed.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Schroder CM, Primeau MM, Hallmayer JF, Lazzeroni LC, Hubbard JT, O’Hara R. Serotonin transporter polymorphism is associated with increased apnea–hypopnea index in older adults. Int J Geriatr Psychiatry. 2014;29:227–235. doi: 10.1002/gps.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lima Marson FA, Bertuzzo CS, Secolin R, Ribeiro AF, Ribeiro JD. Genetic interaction of GSH metabolic pathway genes in cystic fibrosis. BMC Med Genet. 2013;14:60. doi: 10.1186/1471-2350-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DK, Jacobson FL, Washko GR, Casaburi R, Make BJ, Crapo JD, Silverman EK, Hersh CP. Clinical and radiographic correlates of hypoxemia and oxygen therapy in the COPDGene study. Respir Med. 2011;105:1211–1221. doi: 10.1016/j.rmed.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vold ML, Aasebø U, Hjalmarsen A, Melbye H. Predictors of oxygen saturation ≤95% in a cross-sectional population based survey. Respir Med. 2012;106:1551–1558. doi: 10.1016/j.rmed.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Bright NJ, Carling D, Thornton C. Investigating the regulation of brain-specific kinases 1 and 2 by phosphorylation. J Biol Chem. 2008;283:14946–14954. doi: 10.1074/jbc.M710381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo S, Matsunaga T, Kitade Y, Ohno S, Tajima K, El-Kabbani O, Hara A. Human carbonyl reductase 4 is a mitochondrial NADPH–dependent quinone reductase. Biochem Biophys Res Commun. 2008;377:1326–1330. doi: 10.1016/j.bbrc.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Cohen MM., Jr Perspectives on RUNX genes: an update. Am J Med Genet A. 2009;149A:2629–2646. doi: 10.1002/ajmg.a.33021. [DOI] [PubMed] [Google Scholar]
- 32.Jagannathan-Bogdan M, Zon LI. Hematopoiesis. Development. 2013;140:2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tijchon E, Havinga J, van Leeuwen FN, Scheijen B. B-lineage transcription factors and cooperating gene lesions required for leukemia development. Leukemia. 2013;27:541–552. doi: 10.1038/leu.2012.293. [DOI] [PubMed] [Google Scholar]
- 34.Shen JT, Falanga V. Growth factors, signal transduction, and cellular responses. J Dermatol. 2003;30:5–16. [PubMed] [Google Scholar]
- 35.Van Kim CL, Colin Y, Cartron JP. Rh proteins: key structural and functional components of the red cell membrane. Blood Rev. 2006;20:93–110. doi: 10.1016/j.blre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Boos D, Yekezare M, Diffley JF. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science. 2013;340:981–984. doi: 10.1126/science.1237448. [DOI] [PubMed] [Google Scholar]
- 37.Coles GL, Ackerman KG. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2013;110:E1898–E1905. doi: 10.1073/pnas.1222797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh Y, Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review) Int J Mol Med. 2006;18:1019–1023. [PubMed] [Google Scholar]
- 39.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim WJ, Oh YM, Lee JH, Park CS, Park SW, Park JS, Lee SD. Genetic variants in HHIP are associated with FEV1 in subjects with chronic obstructive pulmonary disease. Respirology. 2013;18:1202–1209. doi: 10.1111/resp.12139. [DOI] [PubMed] [Google Scholar]
- 41.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper RS, Tayo B, Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet. 2008;17:R151–R155. doi: 10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 44.Simonson TS, McClain DA, Jorde LB, Prchal JT. Genetic determinants of Tibetan high-altitude adaptation. Hum Genet. 2012;131:527–533. doi: 10.1007/s00439-011-1109-3. [DOI] [PubMed] [Google Scholar]
- 45.Beall CM. Human adaptability studies at high altitude: research designs and major concepts during fifty years of discovery. Am J Hum Biol. 2013;25:141–147. doi: 10.1002/ajhb.22355. [DOI] [PubMed] [Google Scholar]
- 46.Aggarwal S, Negi S, Jha P, Singh PK, Stobdan T, Pasha MA, Ghosh S, Agrawal A, Prasher B, Mukerji M Indian Genome Variation Consortium. EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proc Natl Acad Sci USA. 2010;107:18961–18966. doi: 10.1073/pnas.1006108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu, Basang, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 49.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]