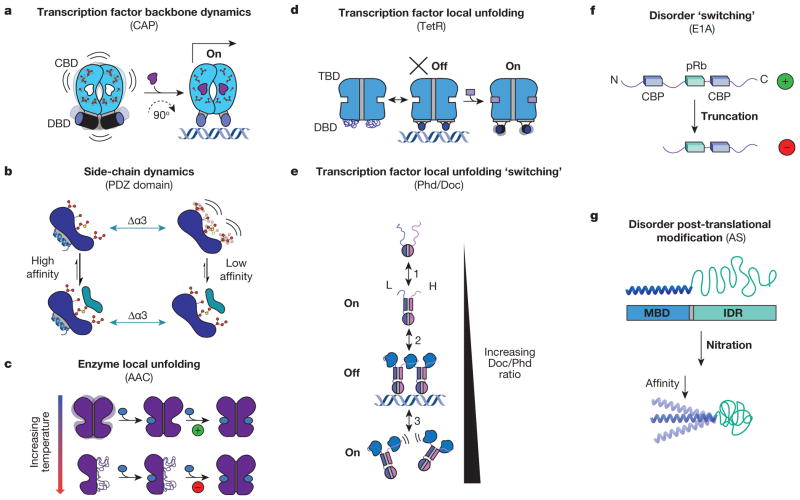
Figure 3. Allosteric systems from the dynamic continuum.
a, CAP homodimer with a cyclic nucleotide binding domain (CBD, blue domains with side chains) and a DNA binding domain (DBD, blue cylinders). Binding energetics of cAMP (purple ligand) quench dynamics in the bound-state ensemble and induce a 90° change in the conformation of the DBD, allowing it to bind DNA and turn on transcription24–26. b, Side-chain dynamics modulate the binding affinity of a canonical PDZ domain to its native ligand with and without α-helix 3 (Δα3, green arrows)23. c, AAC homodimer with each monomer represented as one purple domain. Binding of the allosteric effector acetyl-CoA (blue oval) is positively cooperative at low temperatures (green ‘+’), and negatively cooperative at high temperatures (red ‘−’)4. d, TetR homodimer27 depicted as a two-domain protein with a tetracycline binding domain (TBD, blue region, top) and a DBD (blue region, bottom). e, Doc/Phd toxin–antitoxin system equilibrium. Phd is depicted as a homodimer (top, blue and purple monomers) and Doc is depicted as the blue ligand28. f, Representation of variants of E1A30 shown from N to C terminus with binding sites for ligands in the two variants represented by blue rectangles (CBP) and green rectangles (pRb). g, Schematic representation of α-synuclein (AS) with its N-terminal membrane-binding domain (MBD) coupled to its C-terminal IDR. Upon oxidative stress (nitration) the affinity of the MBD decreases (bottom)29.