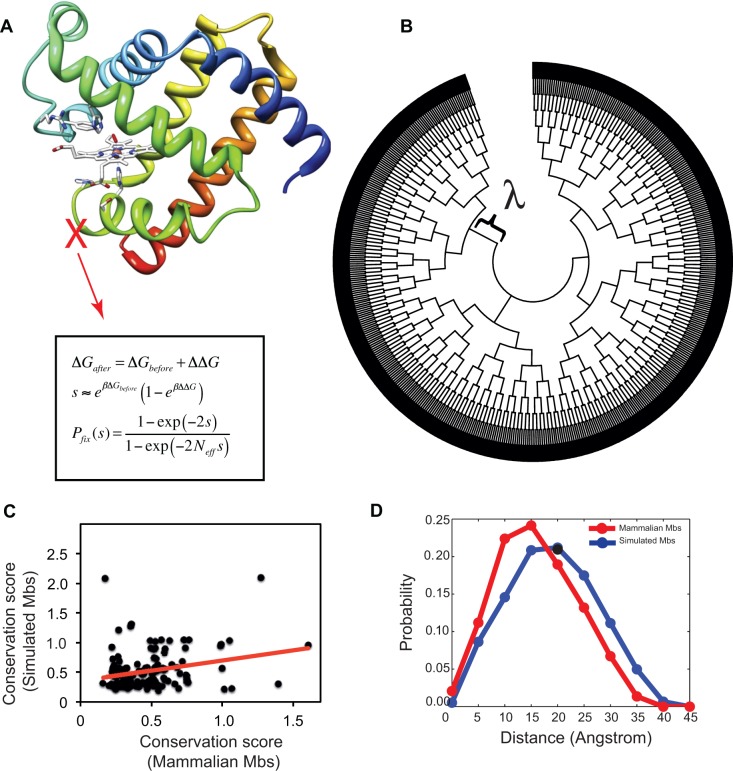
Fig. 1.—
Schematic and performance of structural and evolutionary analyses used in this study. (A) The Mb sequences were evolved in a population of Neff = 104 cells under monoclonal conditions with selection for folding (eqs. 3 and 4). (B) A bifurcating simulated phylogeny with 1,024 external nodes was constructed from an initial Mb sequence with ΔG = −6.84 kcal/mol. Each bifurcation happens after λ arising mutations in the ancestral sequence. (C) Sequence conservation of simulated Mb sequences calculated with Kullback–Leibler score correlates with mammalian Mbs (see Materials and Methods section and supplementary information, Supplementary Material online). (D) The pairwise distance distribution of subsequent substitutions on branches of simulated (in blue) and real mammalian (in red) phylogenetic trees.