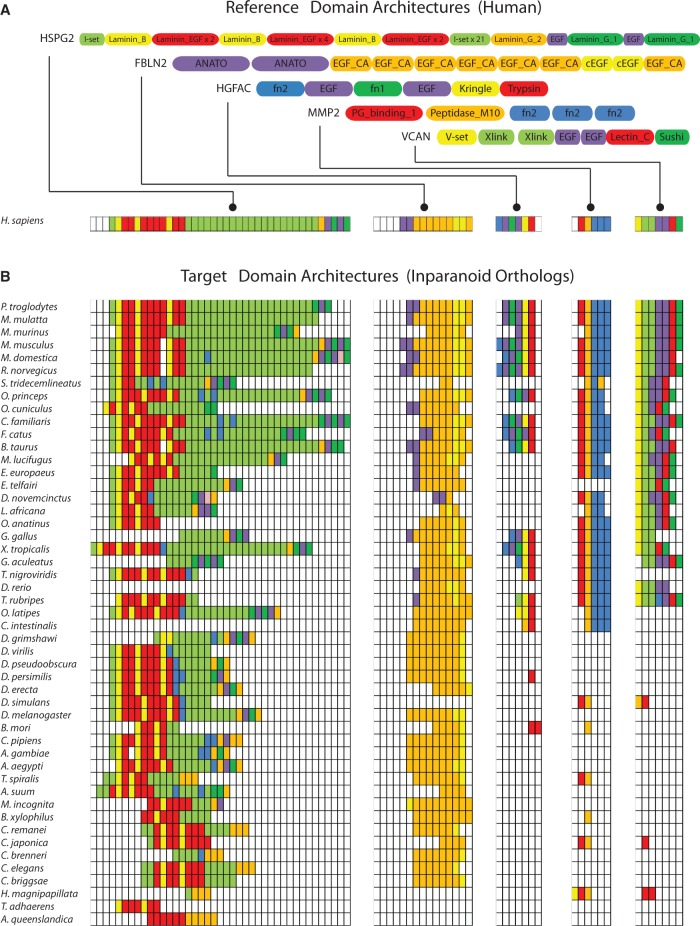
Fig. 3.—
Sample domain architectures illustrating their relative conservation across 131 species with gains and losses of domains occurring throughout. (A) Domain arrangements of human HSPG2, FBLN2, HGFAC, MMP2, and VCAN based on Pfam-A. (B) Corresponding domain based alignments of homologous proteins as detected using Inparanoid. Homologs are arranged in phylogenetic order with mammals at the top. Species with no detectable ortholog are not shown. Full domain alignments for each protein are shown in supplementary spreadsheets S13–S17, Supplementary Material online.