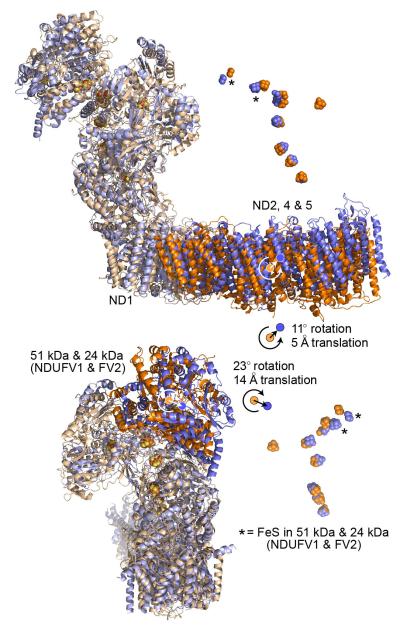
Extended Data Fig. 4. Global comparison of the core subunit structures of bacterial and mammalian complex I.
The core subunits from B. taurus are in blue, and from T. thermophilus (4HEA.pdb4) in orange. The structures have been superimposed using ND1 (the ‘heel’ subunit). Top: the ND2, ND4 and ND5 domain is rotated in B. taurus relative to in T. thermophilus, increasing the curvature in the B. taurus membrane domain. The complex is viewed along the 11° rotation vector (orange) that maps the T. thermophilus ND2, ND4 and ND5 domain to the B. taurus domain, along with a small 5 Å translation to superimpose the domain centres. Correspondingly, the ND3, ND4L and ND6 domains are superimposed by a 4° rotation and a 1 Å translation. Rotation of ND2, 4 and 5 about the long axis of the domain, as noted for Y. lipolytica58, is not observed. Bottom: the NADH dehydrogenase domain containing the 51 and 24 kDa subunits is rotated by 23° and translated by 14 Å in B. taurus, relative to in T. thermophilus, causing the FeS chains to diverge as the distance from ND1 increases. A similar rotation was observed in Y. lipolytica58. The complex is viewed from behind ND1. Correspondingly, the 49 kDa, PSST and TYKY subunits are superimposed by a 6° rotation and a 2 Å translation. The structures were analysed using Superpose from the CCP4 suite59 and the 75 kDa and 30 kDa subunits were not included due to their lower structural conservation.