Figure 3. Minigene Polyadenylation Reporter Analysis.
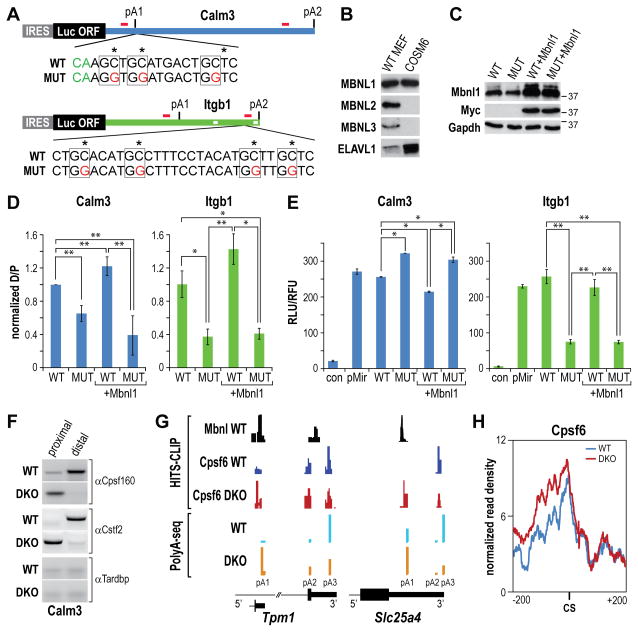
(A) Minigene reporters composed of the Calm3 and Itgb1 3′UTRs (thick green lines) with the proximal (pA1) and distal (pA2) polyadenylation sites cloned downstream of the IRES-driven (grey box) luciferase coding region (turquoise box). The primer binding sites for RT-PCR and 3′ RACE (red bars), the wild type (WT) and mutant (MUT, red letters) sequences downstream (Calm3) or upstream (Itgb1) of the cleavage sites (green CA indicated for Calm3) and the AU-rich binding sites for ELAVL1/HuR (white boxes in Itgb1 3′ UTR) are also indicated.
(B) Relative protein levels for MBNL1, MBNL2, MBNL3 and ELAVL1/HuR in WT MEFs compared to COSM6 cells were determined by immunoblotting.
(C) Immunoblots showing myc-tagged Mbnl1 overexpression following transfection of COSM6 cells with pcDNA3.1-Mbnl1myc. Antibodies to Mbnl1, Myc and Gapdh (loading control) are shown for cells transfected with Calm3WT (WT) and Calm3MUT (MUT) polyadenylation reporters without/with co-transfection with pcDNA3.1-Mbnl1myc (+Mbnl1).
(D) Mbnl proteins repress Calm3 proximal pA (pA1), and activate Itgb1 distal pA (pA2), site use. The ratio of distal to proximal pA selection (D/P) was determined by qRT-PCR for COSM6 cells transfected with the Calm3WT (WT), Calm3MUT (MUT), Itgb13WT (WT), Itgb1MUT (MUT) reporters without/with Mbnl1myc overexpression (+Mbnl1) (data represented as mean +/- SEM, *p < 0.05, **p < 0.01).
(E) Elevated Calm3 pA1 site use leads to enhanced, while loss of Itgb1 distal (pA2) results in decreased, luciferase activity. COSM6 cells were mock transfected (con) or transfected as described in (D) (data represented as mean +/- SEM, *p < 0.05, **p < 0.01).
(F) RNA immunoprecipitation assay with anti-Cpsf160, anti-Cstf2 and anti-Tardbp (control) antibodies.
(G) Examples (Tpm1, Slc25a4) of HITS-CLIP (top) and PolyA-Seq (bottom) showing overlap of Mbnl and Cpsf6 binding sites in wild type (WT), but not Mbnl DKO, MEFs. Enhanced Cpsf6 binding and activation of upstream pA sites were observed following loss of Mbnl activity in DKO MEFs.
(H) RNA map of Cpsf6 normalized CLIP tag density (± 200 bp from the cleavage site, CS) in WT versus DKO MEFs. Polyadenylation sites without additional proximal (±300 bp) sites were selected to avoid interference.