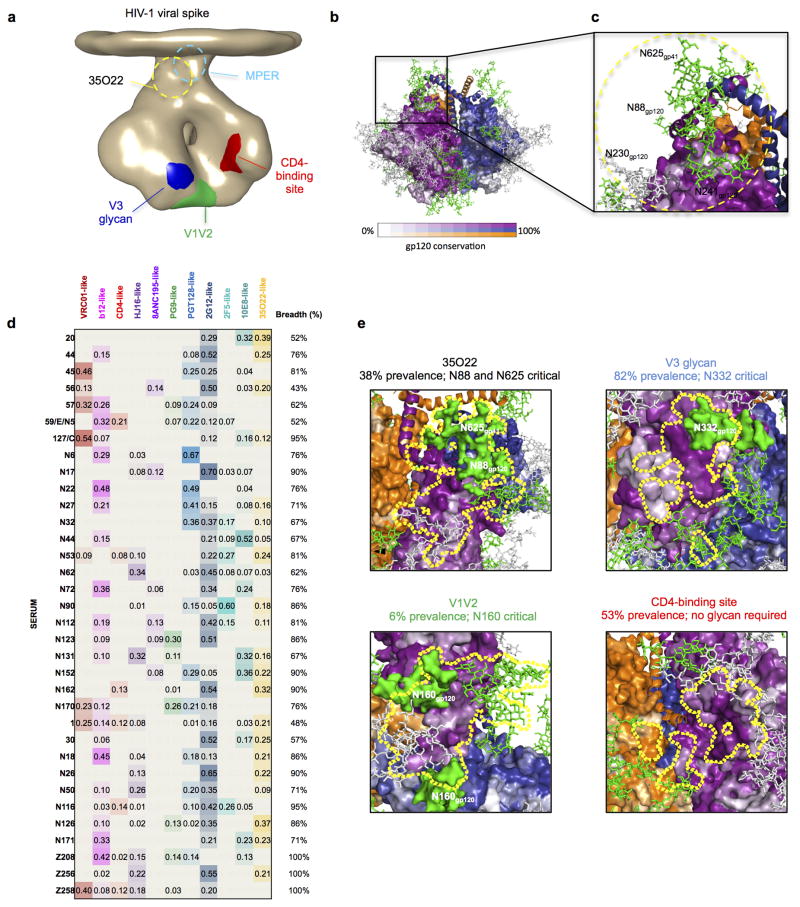
Extended Data Figure 8. A new site of HIV-1 vulnerability at the interface of gp120 and gp41 and prevalence of targeting.
a, Dominant sites of vulnerability to neutralizing antibody elicited by natural infection, shown in the context of an EM tomogram from the BAL viral spike. The viral membrane is positioned at the top of the spike. It is unclear if 35O22 and MPER antibodies bind to this form of the viral spike, and approximate locations for these are shown in dotted outlines. b, Viral spike from the soluble BG505 SOSIP context, shown in the same orientation as a, with gp120 surface colored by conservation from 0–100%, from 4265 HIV-1 strains (white to purple for protomer 1 with scale shown, white to blue for protomer 2 and white to orange for protomer 3), with glycans shown in green when present in more than 90% of strains, in grey when present in 30–90% of strains and not shown otherwise. c, 35O22-identified site of HIV-1 vulnerability, comprises both conserved amino acids and a cluster of glycans, including N88 from gp120 and N625 from gp41. N230 and N241 are not present in BG505 strain. The 35O22 epitope is shown in yellow dotted line. d, Neutralization fingerprints for 35O22 and for antibodies encompassing ten different epitope specificities representing the other four known major sites of Env vulnerability were used to interrogate the serum specificities of 34 HIV-infected patients. Values (with proportional color intensities) predict the fraction of serum neutralization that can be attributed to each antibody specificity. Possible 35O22-like signals were predicted for 13 of the sera (values >0.2), while strong signals were observed in 3 of the sera (values >0.3). A panel of 21 HIV-1 strains was used in the neutralization analysis and for computing serum breadth. e, Sites of HIV-1 vulnerability to neutralizing antibody outlined by a yellow line. Prevalence in 34-donor cohort indicated along with critical glycans.