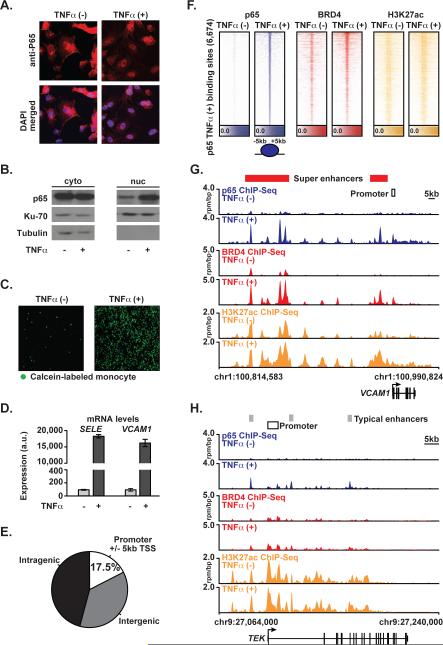
Figure 1. p65 and BRD4 Genome Binding During Proinflammatory Activation in ECs.
(A) Images of ECs ± TNFα stained for p65 (red) or DAPI (blue) (25 ng/mL, 1 hr) cells. (B) Western blot for p65, Ku-70, and Tubulin in cytosolic (left) and nuclear (right) protein fraction lysates in ECs±TNFα. (C) Images showing adhesion of calcein-labeled THP-1 monocytes to ECs ± TNFα (25 ng/mL, 3 hrs). (D) Bar plots showing cell count normalized expression levels of SELE and VCAM1 in ECs±TNFα (25 ng/mL, 3 hrs). Error bars are standard error of the mean (SEM). (E) Pie chart of p65 binding site distribution in EC genome in TNFα(+). (F) Heatmap of p65 (blue), BRD4 (red) and H3K27ac (yellow) levels in resting ECs and after TNFα (25 ng/mL, 1 hr). Each row shows ± 5kb centered on p65 peak. Rows are ordered by max p65 in each region. ChIP-Seq signal (rpm/bp) is depicted by color scaled intensities. (G,H) Gene tracks of ChIP-Seq signal for p65, BRD4, and H3K27ac at the VCAM1 and TEK gene loci in untreated (top) or TNFα(+) (bottom) ECs. Y-axis shows ChIP-Seq signal (rpm/bp). The x-axis depicts genomic position with TNFα gained typical enhancers (TE, gray) and SEs (SE, red) and promoter regions (white) marked. See also Figure S1.