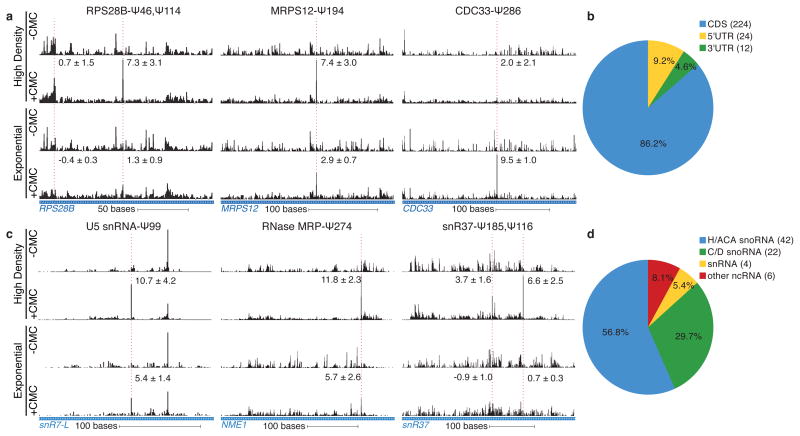
Figure 2. Yeast mRNAs and ncRNAs are inducibly pseudouridylated.
a,c) CMC-dependent peaks of reads are indicated with a dashed red line. The median Pseudo-seq peak heights in each condition are given ±SD, negative peak values occur when the reads in the −CMC library exceed those in the +CMC library. Traces are representative of four wild-type biological replicates. a) Pseudo-seq reads in RPS28B (chrXII:673163–673336), MRPS12 (chrXIV:694489–694736), and CDC33 (chrXV:50560–60875). b) Summary of locations of Ψs within mRNA features. c) Pseudo-seq reads in U5 snRNA (snR7-L, chrVII:939458–939671), RNase MRP RNA (NME1, chrXIV:585585–585925), and an H/ACA snoRNA (snR37, chrX:228090–228475). d) Summary of novel Ψs identified in ncRNA.