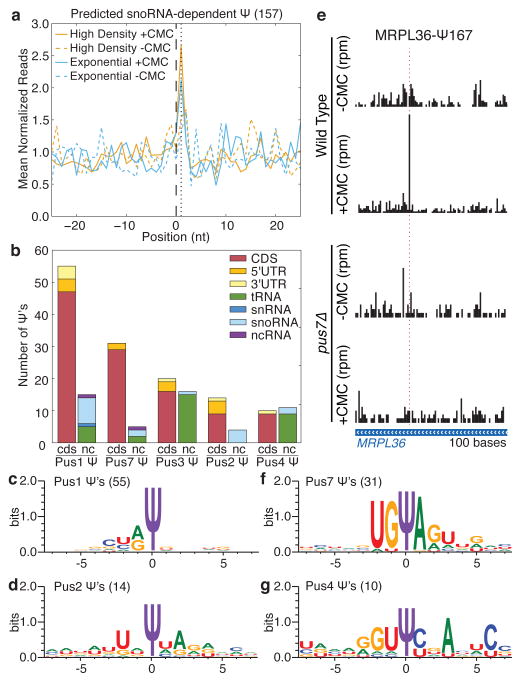
Figure 3. Mechanisms of mRNA pseudouridylation.
a) MetaPsi plot of mean normalized reads for computationally predicted snoRNA-dependent targets in mRNA from high density cultures (orange), log phase cultures (blue), +CMC (solid), and −CMC (dashed). Indicated are the predicted snoRNA target site (black, dashed), and the expected peak of CMC-dependent reads (black, dotted). Reads were pooled from four wild-type biological replicate libraries. b) Summary of Ψs identified by Pseudo-seq as PUS-dependent. The few PUS6- and PUS9-dependent Ψs are not shown. The locations of Ψs within mRNAs and the distribution of Ψs among ncRNA types are indicated. c,d,f,g) Sequence motifs surrounding PUS1- (c), PUS2- (d), PUS7- (f), and PUS4-dependent (g) Ψs in mRNAs, generated by WebLogo 3.3. d) PUS7-dependent Pseudo-seq reads in MRPL36 (chrII:484301–484400).