Abstract
The lysophosphatidic acid receptor 1 (LPA1), a G-protein coupled receptor, regulates cell proliferation, migration, and cytokine release. Here, we investigate the molecular signature of LPA1 trafficking to the cell surface. The overexpressed LPA1 with a C-terminal V5 tag (LPA1-V5) is majorly expressed on the cell surface, while two deletion mutants (C320 and Δ84–87) failed to be trafficked to the cell surface. Further, site-directed mutagenesis analysis of the LPA1 revealed that Ile325, Tyr85, and Leu87 within these two fragments regulate LPA1 maturation and trafficking to the cell surface. Over-expression of Sar1, a component of coat protein complex II (COPII), enhances glycosylation of LPA1 wild type, but not these mutants. The mutants of LPA1 are majorly localized in the endoplasmic reticulum (ER) and exhibit a higher binding affinity to heat shock protein 70 (Hsp70), when compared to the LPA1 wild type. Further, we found that all these mutants failed to increase phosphorylation of Erk, and the cytokine release in response to LPA treatment. These results suggest that Ile325, Tyr85, and Leu87 within LPA1 are essential for LPA1protein properly folding in the ER.
Keywords: GPCR, Cell Surface trafficking, ER quality control, mutation
1. Introduction
Lysophosphatidic acid (LPA) is a naturally occurring bioactive phospholipid that triggers intracellular signals including the increase of intracellular calcium, and the activation of protein kinases and transcriptional factors [1–4]. The biological effects of LPA are dependent on LPA receptors on cell surface, which are member of G-protein coupled receptors (GPCRs) [5]. Among them, LPA receptor 1 (LPA1) is highly expressed in the most cell types including lung epithelial cells [6–8]. LPA1 is considered to be a pro-inflammatory receptor [9–11]. LPA1 deficient mice reduced intratracheal instillation of lipopolysaccharide (LPS)- or bleomycininduced lung injury [9, 11]. LPA1-mediated signals regulate tumor cell migration and proliferation and contribute to the tumorigenesis [12, 13]. LPA1 is a potential target in the development of new therapeutic strategies to lessen lung inflammation and reduce tumor growth and metastasis [9, 10, 14–19].
GPCRs belong to the seven transmembrane receptors which are localized on the cell surface. The expression levels of GPCRs on the cell surface determine the effects of ligand-induced cellular responses. Receptors are synthesized on the rough endoplasmic reticulum (ER). Receptors trafficking from the ER / Golgi to the cell surface regulate the GPCR levels on the cell surface. Once the properly folded receptor has passed the ER quality control, it is transported to the Golgi for glycosylation and maturation [20]. Sar1, a member of coat protein complex II (COPII), is known to regulate protein trafficking from the ER to the Golgi [21, 22]. Misfolded receptors within the ER are recognized by heat-shock protein 70 (Hsp70) family members and destroyed by the ER-associated degradation pathway (ERAD) [23–25].
The PDZ protein GIPC (GAIP-interacting protein, C terminus) promotes LPA1 internalization and attenuates LPA responses [26]. However, the regulation of LPA1 trafficking to cell surface has not been studied. Here, we show three amino acids play a vital role in the regulation of LPA1 glycosylation, trafficking to the cell surface, and LPA1-mediated signaling.
2. Materials and methods
2.1. Cell culture and reagents
Murine lung epithelial cells (MLE12) and human bronchial epithelial cell (Beas2B) [American Type Culture Collection (ATCC), Manassas, VA, USA] were cultured with HITES medium containing 10 % FBS. All the cells were cultured at 37°C in 5% CO2. V5 antibody, mammalian expressional plasmid pcDNA3.1D/His-V5 TOPO, Escherichia coli Top 10 competent cells were from Invitrogen (Carlsbad, CA, USA). β-actin and Flag antibodies and LPA were from Sigma Aldrich (St. Louis, MO, USA). Hsp70 antibody was obtained from StressMarq Bioscience (Victoria, Canada). Immunobilized protein A/G beads were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All materials in highest grades uses in the experiments are commercially available.
2.2. DNA construction
Human LPA1 cDNA was inserted into pcDNA3.1D/His-V5 TOPO vector. C-terminal or intracellular domain deletion mutants of LPA1 were generated by PCR with specific primers designed to target LPA1 cDNA sequence. Site directed mutagenesis was performed to generate LPA1 mutants according to the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA).
2.3. Immunoblotting and immunoprecipitation
Cells were washed with cold PBS and collected in cell lysis buffer. An equal amount of cell lysates (20 µg) was subjected to SDS-PAGE without boiling, electrotransferred to membranes and immunoblotted as standard protocol. For immunoprecipitation, equal amounts of cell lysates (1 mg) were incubated with a V5 tag antibody overnight at 4 °C, followed by the addition of 40 µl of protein A/G agarose and incubation for additional 2 h at 4 °C. The immunoprecipitated complex was washed 3 times with 1% Triton X-100 in ice cold phosphate-buffered saline and analyzed by immunoblotting with an Hsp70 antibody.
2.4. Immunostaining
MLE12 cells were plated on 35 mm glass-bottom culture dishes. After LPA treatment, cells were fixed in 3.7% formaldehyde for 20 min, followed by permeabilization with 0.1% Triton X-100 for 2 min. Cells were incubated with 1:200 dilution of V5 tag antibody, followed by a 1:200 dilution of fluorescence-conjugated secondary antibody sequentially for immunostaining.
2.5. IL-8 Elisa assay
Beas2B cells were treated with LPA for 3 h, and then supernatants were collected. Human IL-8 levels in the supernatants were measured by IL-8 Elisa kit according to the manufacturer’s instruction.
3. Results and discussion
3.1. Ile325 within LPA1 regulates LPA1 trafficking to the cell surface and maturation
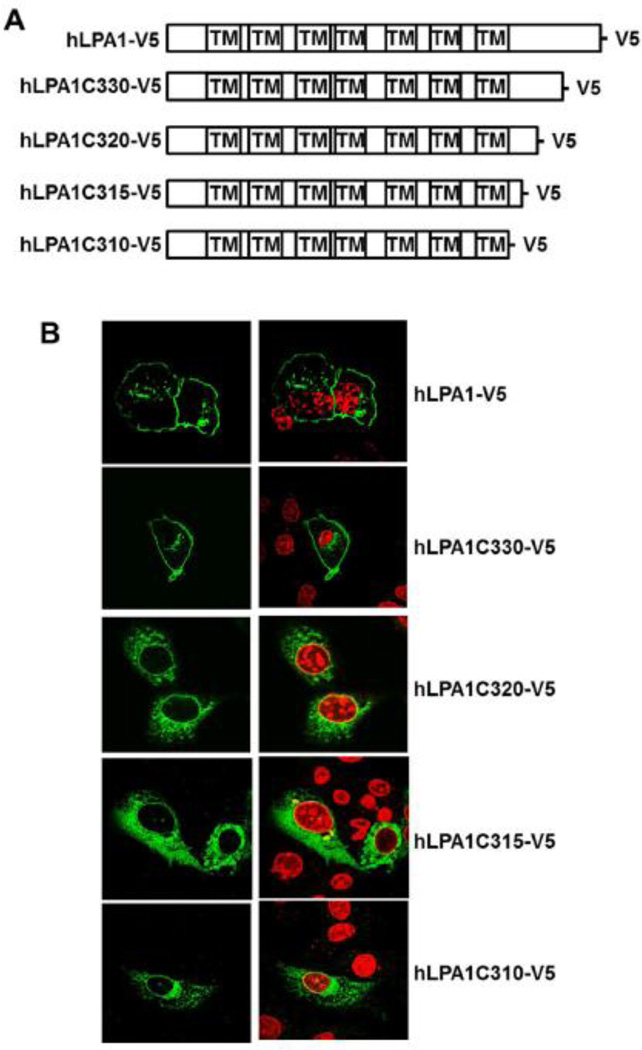
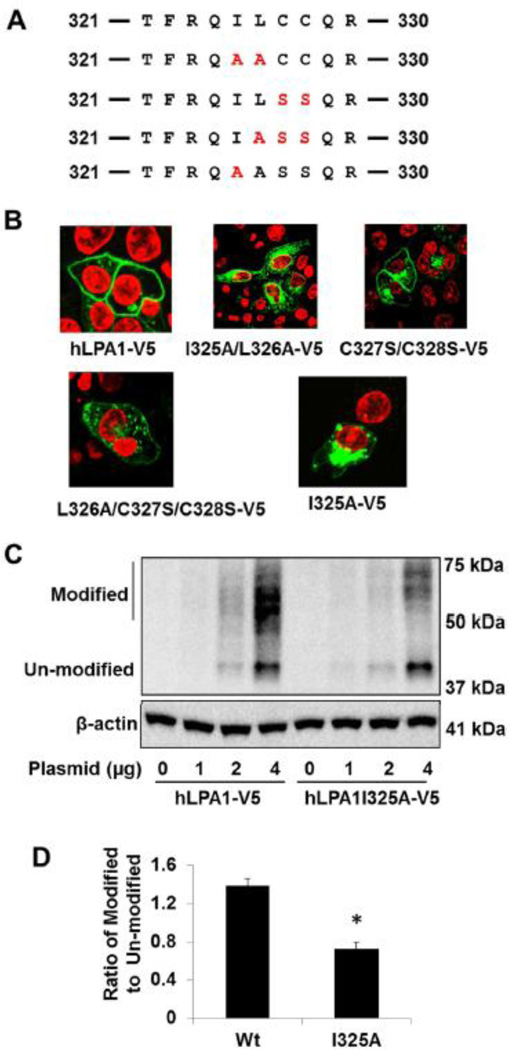
Modulation of receptor trafficking to the cell surface regulates the receptor-mediated signals and biological functions. LPA1 is a receptor with multiple functions, belonging to the GPCR family. GPCR export to the cell surface membrane is tightly regulated by the ER quality-control system, specific protein partners, and post-translational modifications. To investigate which fragment of LPA1 regulates its intracellular trafficking, we generated plasmids of LPA1 wild type and C-terminal deletion mutants with a V5 tag in the C-termini. Immunostaining with a V5 tag antibody revealed that LPA1-V5 and c-terminal deletion mutant (LPA1C330-V5) are localized primarily on the cell surface, however, further deletion of LPA1 (LPA1C320-V5, LPA1C315-V5, and LPA1C310-V5) failed to traffic to the cell surface (Fig. 1A and 1B). This data indicates that amino acids between 320 and 330 play a vital role in the regulation of LPA1 trafficking. To identify which amino acid is involved, several amino acids between 320 and 330 were mutated as shown in Fig. 2A. Unlike other LPA1 mutants, mutants containing Ile 325 (LPA1I325A-V5 and LPA1I325A/L326A-V5) are majorly expressed inside the cytoplasm (Fig. 2B), suggesting that Ile325 is a key amino acid in the regulation of LPA1 trafficking. Ile325 is in the dileucine like region, which has been known to regulate receptor trafficking. The dileucine motif in the vasopressin receptor regulates its trafficking from ER to the cell surface [27, 28], while the dileucine motif of oncostatin M receptor mediates the receptor trafficking from the cell surface to the cytoplasm [29]. Here, we show that the dileucine like region within LPA1, plays a crucial role in the escaping the receptor from ER to the cell surface. There is double cysteine residues (C327/C328) in the fragment, which is considered as a canonical palmitoylation motif [30, 31]. We found the double cysteine residues mutation (LPA1C327S/C328S) had no effect on the alteration of LPA1 expression on the cell surface. Post-translational modification regulates GPCRs trafficking to the cell surface. SDS-PAGE gel analysis with a V5 antibody shows that LPA1-V5 has more modified forms (50–70 kDa) than un-modified forms (40 kDa), while LPA1I325A-V5 has less modified forms that the un-modified forms (Fig. 2C and 2D). These data indicate that the LPA1I325A-V5 mutant fails to be post-translational modified and is unable to move to the cell surface.
Figure 1. Residues 320–330 are involved in LPA1 trafficking to the cell surface.
A. LPA1 wild type and c-terminal deletion mutants were inserted into pCDNA3.1/Topo-V5-His plasmids. B. Ectopic expressed LPA1 wild type and c-terminal deletion mutants with V5 tag in MLE12 cells were immunostained with a V5 antibody (green). Nuclei were stained with DAPI (red).
Figure 2. Ile325 regulates LPA1 trafficking to the cell surface.
A. Sequence of amino acids between 321 to 330 within LPA1. Site mutations are shown in red. B. LPA1 wild type and point mutants with V5 tag were expressed in MLE12 cells and immunostained with a V5 antibody (green). Nuclei were stained with DAPI (red). C. MLE12 cells were transfected with LPA1-V5 and LPA1I325A-V5 plasmids (0–4 µg) and cell lysates were analyzed with immunoblotting with antibodies to V5 tag and β-actin. D. Intensity ratio of modified to un-modified in 4 µg plasmid transfected cells was quantified. *p<0.01, compared to LPA1 wild type (Wt). All the images and blots were representative from three independent experiments.
3.2. Tyr85 and Leu87 are critical in the regulation of LPA1 trafficking to the cell surface
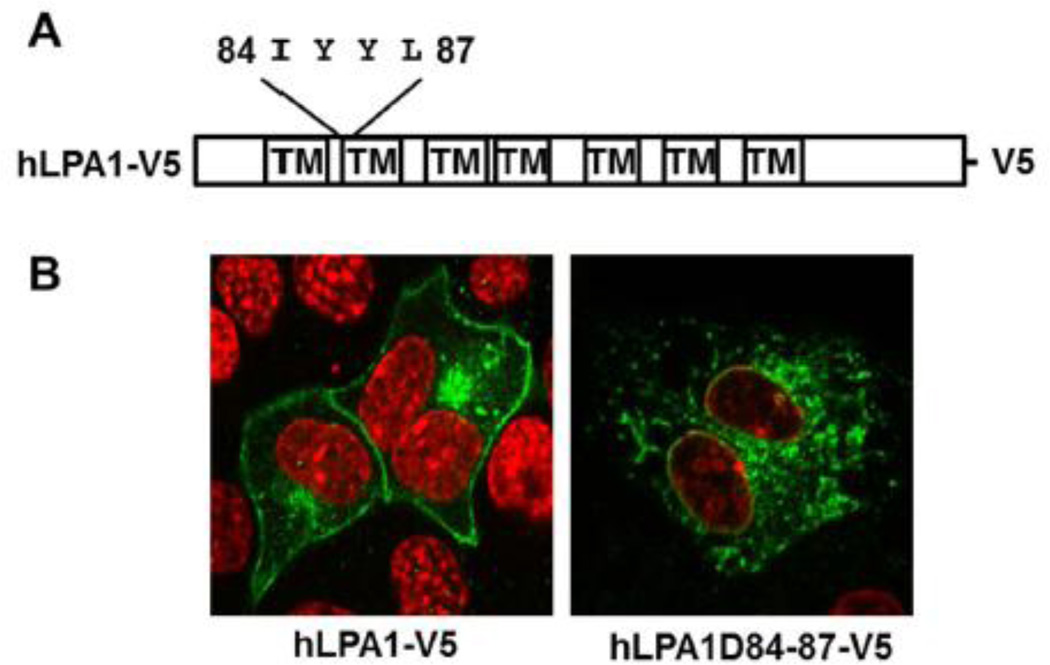
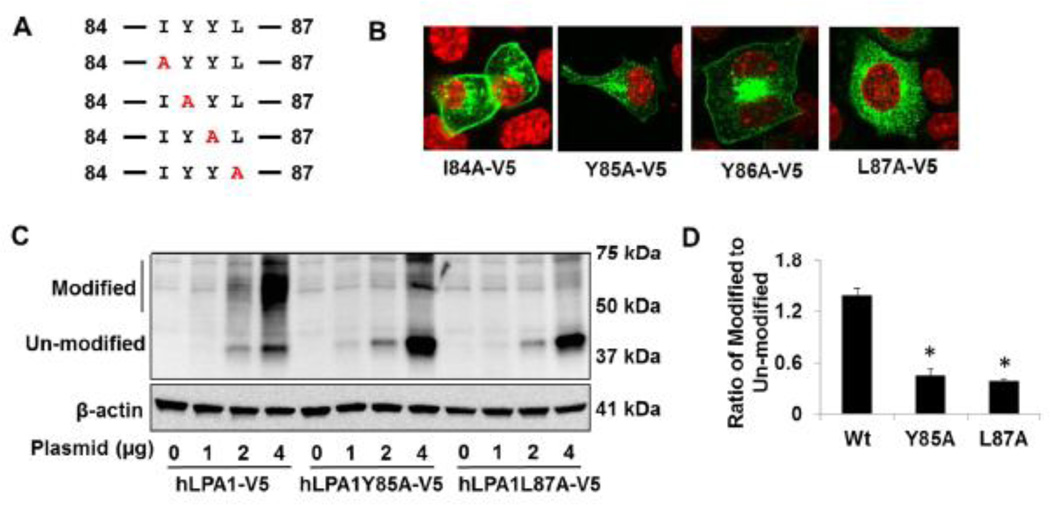
In addition to the c-termini of LPA1, we also found that deletion of four amino acids (84–87, IYYL) within the first intracellular loop results in failure of LPA1 trafficking to the cell surface (Fig. 3A and3 B). Analysis of a series of point mutants reveals that LPA1Y85A-V5 and LPA1L87A-V5, not LPA1I84A-V5 or LPA1L87A loss their ability to express on the cell surface (Fig. 4A and 4B). Further, these two mutants exhibit less modification when compared to wild type LPA1-V5 (Fig. 4C and 4D). These data suggest that both Tyr85 and Leu87 within the first intracellular loop are important for LPA1 post-translational modification and trafficking to the cell surface. Membrane proximal tyrosine residue in Vpu protein from HIV- subtype C has been known to regulate the protein transporting to the cell surface [32]. We found Tyr85, but not Tyr86, contributes to the LPA1 trafficking. The tyrosine residue is an acceptor site for phosphorylation and plays a role in protein-protein interaction. The Cardiac sodium channel binding to calcium binding protein calmodulin is dependent on the double tyrosine motif [33]. The experimental evidence for phosphorylation at Tyr85 in LPA1 has not been revealed. It may be a site for LPA1 interaction with chaperone proteins during protein synthesis and folding.
Figure 3. Residues 84–87 are involved in LPA1 trafficking to the cell surface.
A. The LPA1 structure with highlighted residues 84–87. B. Ectopically expressed LPA1 wild type and amino acids 84–87 deletion mutants with V5 tag in MLE12 cells were immunostained with a V5 antibody (green). Nuclei were stained with DAPI (red).
Figure 4. Tyr85 and Leu87 regulates LPA1 trafficking to the cell surface.
A. Sequence of amino acids between 84 to 87 within LPA1. Site mutations were shown in red. B. LPA1 wild type and point mutants with V5 tag were expressed in MLE12 cells and immunostained with a V5 antibody (green). Nuclei were stained with DAPI (red). C. MLE12 cells were transfected with LPA1-V5, LPA1Y85A-V5, and LPA1L87A-V5 plasmids (0–4 µg) and cell lysates were then analyzed with immunoblotting with antibodies to V5 tag and β-actin. D. Intensity ratio of modified to un-modified in 4 µg plasmid transfected cells was quantified. *p<0.01, compared to LPA1 wild type (Wt). All the images and blots were representative from three independent experiments.
3.3. LPA1I325A, LPA1Y85A, and LPA1L87A mutants accumulate within the ER affecting LPA1 glycosylation
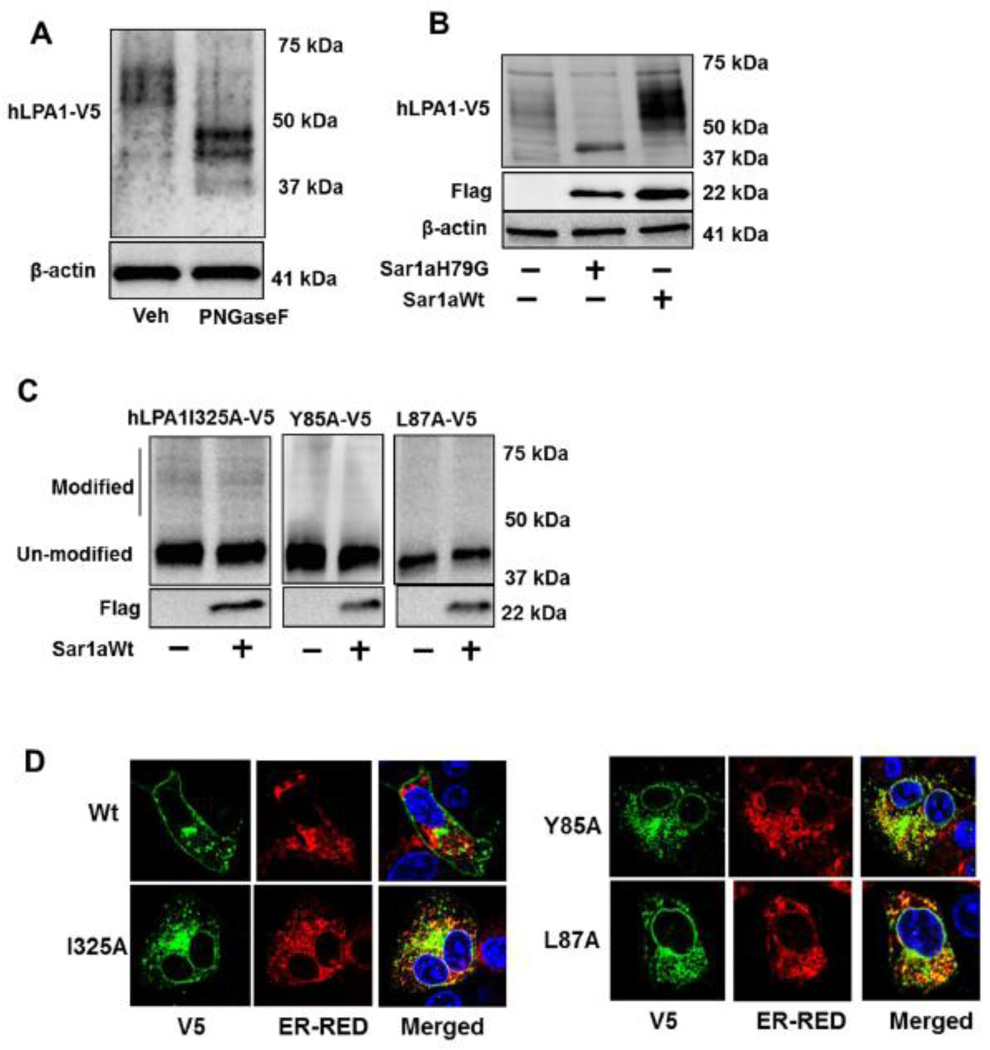
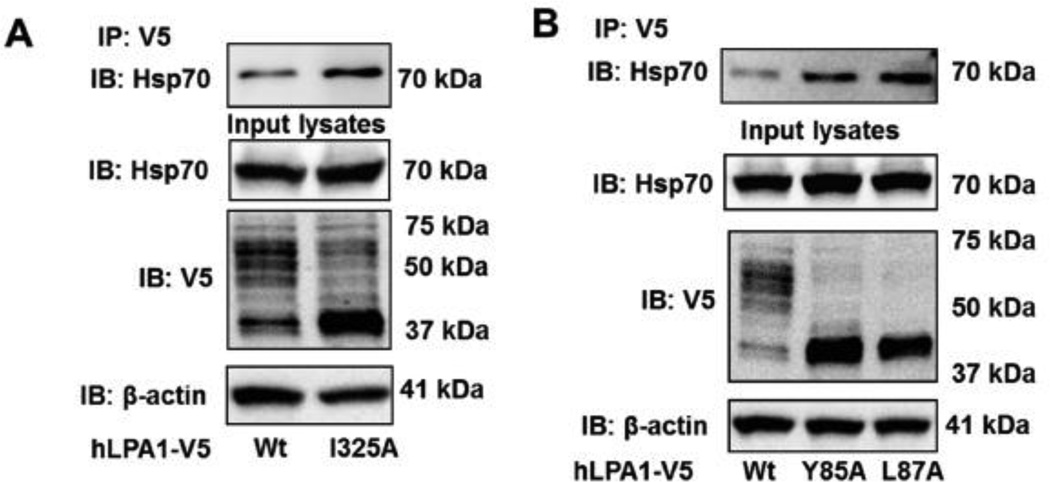
N-glycosylation regulates GPCR structure and maturation [34–36]. Consistent with the findings from Versano et al. [26], a prominent broad band of LPA1-V5 at ~60 kDa was shifted to ~40 kDa after treatment with PNGase F, an endoglycosidase to remove N-linked glycans (Fig. 5A). Receptor glycosylation is catalyzed either in the ER or Golgi. Sar1, a small GTPase, has been identified to promote vesicle budding from the ER and mediates protein trafficking from the ER to Golgi [22]. Sar1 is a component of coat protein complex II (COPII), which is formed on the ER membrane at ER exit sites [37]. LPA1 glycosylation was enhanced by over-expression of Sar1a wild type, and was attenuated by dominant negative Sar1aH79G (Fig. 5B). However, the Sar1a wild type failed to promote the glycosylation of LPA1I325A, LPA1Y85A, and LPA1L87A mutants (Fig. 5C). This suggests that Sar1 promotes LPA1 from the ER to Golgi but is unable to promote similar actions for the site directed mutants. This finding is consistent with the studies that describe Sar1 regulates cell surface expression of GPCRs including At1R, β2- AR, and α26-AR [38]. The data lead us to hypothesize that these mutants failed to traffic from the ER to the Golgi, where LPA1 is N-linked glycosylated. The data from co-immunostaining with V5 antibody and ER marker indicates that all these mutants, but not LPA1 wild type, are localized in the ER (Fig. 5D). N-linked glycosylation usually occurs on the N-terminal or the extracellular loop of the receptors. Since none of these amino acids are considered receptors for N-linked glycans, we hypothesized that these mutants are improperly folded proteins recognized by ER quality control system, which includes cytoplasmic heat shock protein 70 (Hsp70). Hsp70 binds to improperly folded proteins but not their native conformation [39]. We found that all these LPA1 mutants enhanced the association with Hsp70, when compared to LPA1 wild type (Fig. 6A and 6B), suggesting that these single amino acid mutations cause LPA1 conformational changes, misfolding, and failure to traffic to the Golgi and cell surface.
Figure 5. LPA1I325A, LPA1Y85A, and LPA1L87A mutants accumulate within the ER.
A. LPA1-V5 over-expressed cell lysates were treated with or without PNGase F, followed by V5 and β-actin immunobloting. B. LPA1-V5 was co-expressed with Sar1aH79G-Flag or Sar1aWt- Flag in MLE12 cells. Cell lysates were analyzed for V5, Flag, and β-actin immunobloting. C. LPA1I325A-V5, LPA1Y85A-V5, or LPA1L87A-V5 was co-overexpressed with Sar1aWt-Flag in MLE12 cells. Cell lysates were analyzed for V5 and Flag immunobloting. D. LPA1 wild type and point mutants with V5 tag were co-overexpressed with DsRed2-ER in MLE12 cells. Immunostaining were performed with a V5 antibody (green). DsRed2-ER (ER-RED) shows in red and nuclei were stained with DAPI (blue).
Figure 6. LPA1I325A, LPA1Y85A, and LPA1L87A mutants interact with Hsp70.
LPA1 wild type- and point mutants with V5 tag- overexpressed MLE12 cell lysates were subjected to immunoprecipitation with a V5 antibody, followed by Hsp70 immunobloting. Input lysates were analyzed with Hsp70, V5, and β-actin antibodies. All the images and blots are representative from three independent experiments.
3.4. LPA1I325A, LPA1Y85A, and LPA1L87A mutants are not activated by LPA
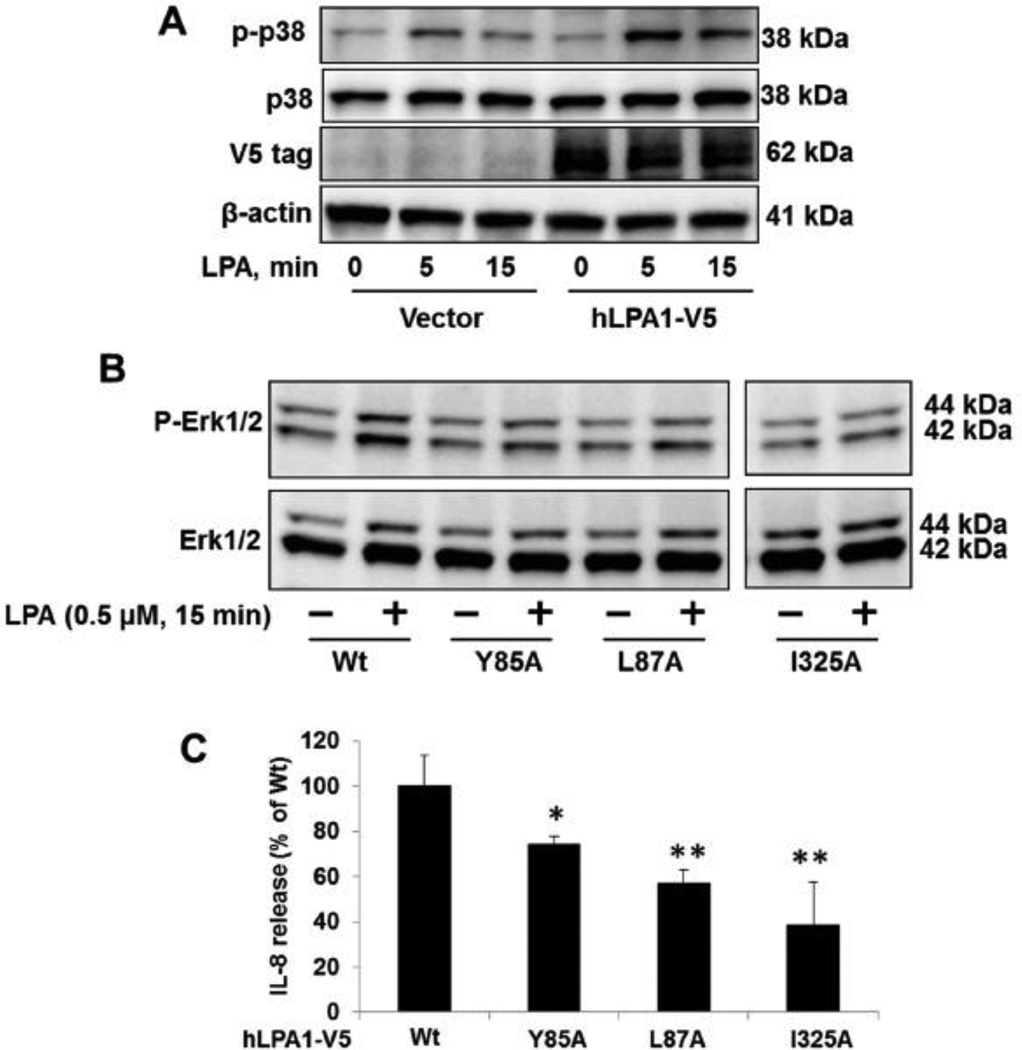
Similar to other cell surface receptors, the biological functions of LPA1 is dependent on ligation to its ligand, LPA. We hypothesized that LPA1 mutants are not activated by LPA due to their accumulation within the ER, not on the cell surface. First, we examined how the over-expressed LPA1-V5 responded to LPA treatment. As shown in Fig. 7A, over-expression of LPA1-V5 enhanced LPA-induced phosphorylation of p38 MAPK [40], suggesting that ectopically expressed LPA1-V5 is the functional LPA1 receptor on the cell surface. Unlike LPA1-V5, overexpression of LPA1Y85A, LPA1L87A, or LPA1I325A did not enhance LPA-induced phosphorylation of Erk1/2 (Fig. 7B). Notably, the 0.5 µM of LPA here has no effect on Erk activation in A549 cells (data not shown). Further, we found that all these mutants were less effective on IL-8 release in response to LPA treatment in human bronchial epithelial cells, when compared to over-expressed LPA1 wild type (Fig. 7C). These data indicate that these mutants are not functional due to their expression failure on the cell surface. LPA1 has been known as a pro-inflammatory factor in lung disorders including lung fibrosis and acute lung injury [9, 10]. LPA1-mediate signals contribute to tumor cell proliferation and migration [12, 13]. To date, there has been no evidence to indicate that these mutants are related to human diseases. The human single nucleotide polymorphism (SNPs) database has been increasing rapidly in recent years. This study provides a fundamental mechanism into the understanding of why these mutants lose their biological functions. Future studies will seek to uncover if these mutants are related to human inflammatory diseases.
Figure 7. Effects of LPA1 mutants on LPA-induced signal.
A. After MLE12 cells were transfected with empty vector or LPA1-V5 plasmid for 48 h, cells were treated with LPA (5 µM) for 5 and 15 min. Cell lysates were analyzed by phosphor (p)-p38, p38, V5 tag, and β-actin. All blots are representative from three independent experiments. B. LPA1 wild type and point mutants over-expressed MLE12 cells were treated with LPA (0.5 µM, 15 min). Cell lysates were analyzed by p-Erk1/2 and Erk1/2. All blots are representative from three independent experiments. C. LPA1 wild type and point mutants over-expressed human bronchial epithelial cells were treated with LPA for 3 h. IL-8 release within medium samples were analyzed by IL-8 Elisa kit. *p<0.05, **p<0.01, when compared to LPA1 wild type (Wt).
Highlights.
Our study identified novel residues within LPA1 which regulate LPA1 maturation and trafficking to the cell surface. This study revealed molecular regulation of GPCR trafficking and provided biochemical information for human diseases SNPs study in the future.
Acknowledgement
This study was supported by the US National Institutes of Health (R01 HL01916 and R01HL112791 to Y.Z.) and American Heart Association awards 12SDG9050005 (J.Z.).
Abbreviations
- LPA
lysophosphatidic acid
- GPCR
G protein-coupled receptor
- ER
endoplasmic reticulum
- Hsp70
heat shock protein 70
- ERAD
ER-associated degradation pathway
- COPII
coat protein complex II.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Xu YJ, Saini HK, Cheema SK, Dhalla NS. Cell Calcium. 2005;38:569–579. doi: 10.1016/j.ceca.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, He D, Stern R, Usatyuk PV, Spannhake EW, Salgia R, Natarajan V. Cellular signalling. 2007;19:2329–2338. doi: 10.1016/j.cellsig.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu SS, Chiu T, Rozengurt E. Am J Physiol Cell Physiol. 2002;282:C1432–C1444. doi: 10.1152/ajpcell.00323.2001. [DOI] [PubMed] [Google Scholar]
- 4.He D, Natarajan V, Stern R, Gorshkova IA, Solway J, Spannhake EW, Zhao Y. The Biochemical journal. 2008;412:153–162. doi: 10.1042/BJ20071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Corven EJ, Groenink A, Jalink K, Eichholtz T, Moolenaar WH. Cell. 1989;59:45–54. doi: 10.1016/0092-8674(89)90868-4. [DOI] [PubMed] [Google Scholar]
- 6.Contos JJ, Ishii I, Chun J. Molecular pharmacology. 2000;58:1188–1196. doi: 10.1124/mol.58.6.1188. [DOI] [PubMed] [Google Scholar]
- 7.Ye X. Hum Reprod Update. 2008;14:519–536. doi: 10.1093/humupd/dmn023. [DOI] [PubMed] [Google Scholar]
- 8.An S, Bleu T, Hallmark OG, Goetzl EJ. The Journal of biological chemistry. 1998;273:7906–7910. doi: 10.1074/jbc.273.14.7906. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, He D, Su Y, Berdyshev E, Chun J, Natarajan V, Zhao Y. Am J Physiol Lung Cell Mol Physiol. 2011;301:L547–L556. doi: 10.1152/ajplung.00058.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaney JS, Chapman C, Correa LD, Stebbins KJ, Bundey RA, Prodanovich PC, Fagan P, Baccei CS, Santini AM, Hutchinson JH, Seiders TJ, Parr TA, Prasit P, Evans JF, Lorrain DS. British journal of pharmacology. 2010;160:1699–1713. doi: 10.1111/j.1476-5381.2010.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tager AM, LaCamera P, Shea BS, Campanella GS, Selman M, Zhao Z, Polosukhin V, Wain J, Karimi-Shah BA, Kim ND, Hart WK, Pardo A, Blackwell TS, Xu Y, Chun J, Luster AD. Nat Med. 2008;14:45–54. doi: 10.1038/nm1685. [DOI] [PubMed] [Google Scholar]
- 12.Alemayehu M, Dragan M, Pape C, Siddiqui I, Sacks DB, Di Guglielmo GM, Babwah AV, Bhattacharya M. PloS one. 2013;8:e56174. doi: 10.1371/journal.pone.0056174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahle M, Veit C, Bachfischer U, Schierling K, Skripczynski B, Hall A, Gierschik P, Giehl K. Journal of cell science. 2003;116:3835–3846. doi: 10.1242/jcs.00679. [DOI] [PubMed] [Google Scholar]
- 14.Boucharaba A, Serre CM, Guglielmi J, Bordet JC, Clezardin P, Peyruchaud O. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9643–9648. doi: 10.1073/pnas.0600979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herr KJ, Herr DR, Lee CW, Noguchi K, Chun J. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15444–15449. doi: 10.1073/pnas.1106129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada T, Sato K, Komachi M, Malchinkhuu E, Tobo M, Kimura T, Kuwabara A, Yanagita Y, Ikeya T, Tanahashi Y, Ogawa T, Ohwada S, Morishita Y, Ohta H, Im DS, Tamoto K, Tomura H, Okajima F. The Journal of biological chemistry. 2004;279:6595–6605. doi: 10.1074/jbc.M308133200. [DOI] [PubMed] [Google Scholar]
- 17.Yamada T, Obo Y, Furukawa M, Hotta M, Yamasaki A, Honoki K, Fukushima N, Tsujiuchi T. Biochemical and biophysical research communications. 2009;378:424–427. doi: 10.1016/j.bbrc.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 18.Obo Y, Yamada T, Furukawa M, Hotta M, Honoki K, Fukushima N, Tsujiuchi T. Mutation research. 2009;660:47–50. doi: 10.1016/j.mrfmmm.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Orosa B, Garcia S, Martinez P, Gonzalez A, Gomez-Reino JJ, Conde C. Annals of the rheumatic diseases. 2014;73:298–305. doi: 10.1136/annrheumdis-2012-202832. [DOI] [PubMed] [Google Scholar]
- 20.Duvernay MT, Filipeanu CM, Wu G. Cellular signalling. 2005;17:1457–1465. doi: 10.1016/j.cellsig.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Pathre P, Shome K, Blumental-Perry A, Bielli A, Haney CJ, Alber S, Watkins SC, Romero G, Aridor M. The EMBO journal. 2003;22:4059–4069. doi: 10.1093/emboj/cdg390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pucadyil TJ, Schmid SL. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolz A, Wolf DH. Biochimica et biophysica acta. 2010;1803:694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Hartl FU, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 25.Meusser B, Hirsch C, Jarosch E, Sommer T. Nature cell biology. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 26.Varsano T, Taupin V, Guo L, Baterina OY, Jr, Farquhar MG. PloS one. 2012;7:e49227. doi: 10.1371/journal.pone.0049227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robert J, Clauser E, Petit PX, Ventura MA. The Journal of biological chemistry. 2005;280:2300–2308. doi: 10.1074/jbc.M410655200. [DOI] [PubMed] [Google Scholar]
- 28.Schulein R, Hermosilla R, Oksche A, Dehe M, Wiesner B, Krause G, Rosenthal W. Molecular pharmacology. 1998;54:525–535. doi: 10.1124/mol.54.3.525. [DOI] [PubMed] [Google Scholar]
- 29.Radtke S, Jorissen A, de Leur HS, Heinrich PC, Behrmann I. The Journal of biological chemistry. 2006;281:4024–4034. doi: 10.1074/jbc.M511779200. [DOI] [PubMed] [Google Scholar]
- 30.Dong X, Mitchell DA, Lobo S, Zhao L, Bartels DJ, Deschenes RJ. Molecular and cellular biology. 2003;23:6574–6584. doi: 10.1128/MCB.23.18.6574-6584.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murph MM, Nguyen GH, Radhakrishna H, Mills GB. Biochimica et biophysica acta. 2008;1781:547–557. doi: 10.1016/j.bbalip.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abada P, Noble B, Cannon PM. Journal of virology. 2005;79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarhan MF, Van Petegem F, Ahern CA. The Journal of biological chemistry. 2009;284:33265–33274. doi: 10.1074/jbc.M109.052910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleizen B, Braakman I. Current opinion in cell biology. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Williams DB. Journal of cell science. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 36.Deslauriers B, Ponce C, Lombard C, Larguier R, Bonnafous JC, Marie J. The Biochemical journal. 1999;339(Pt 2):397–405. [PMC free article] [PubMed] [Google Scholar]
- 37.Blumental-Perry A, Haney CJ, Weixel KM, Watkins SC, Weisz OA, Aridor M. Developmental cell. 2006;11:671–682. doi: 10.1016/j.devcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Dong C, Zhou F, Fugetta EK, Filipeanu CM, Wu G. Cellular signalling. 2008;20:1035–1043. doi: 10.1016/j.cellsig.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kampinga HH, Craig EA. Nature reviews. Molecular cell biology. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saatian B, Zhao Y, He D, Georas SN, Watkins T, Spannhake EW, Natarajan V. The Biochemical journal. 2006;393:657–668. doi: 10.1042/BJ20050791. [DOI] [PMC free article] [PubMed] [Google Scholar]