Figure 1.
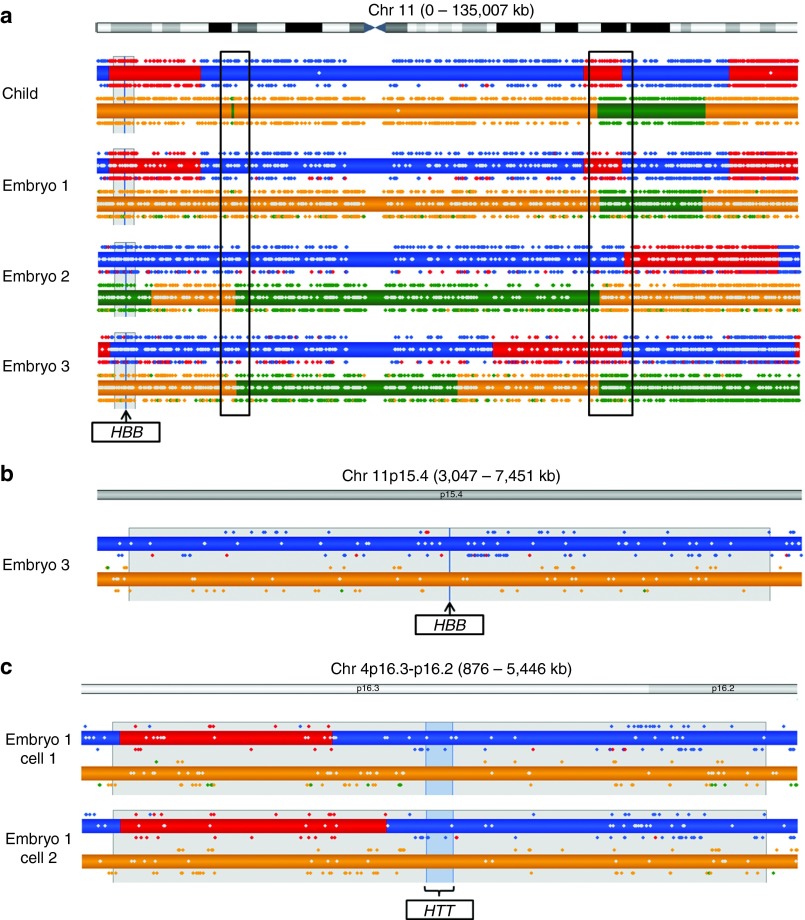
Karyomaps of single blastomeres biopsied from cleavage-stage embryos. Paternal haplotypes are represented in blue/red, and maternal haplotypes are represented in orange/green. Haplotypes inherited by the reference are shown in blue/orange. For a detailed description of how the karyomaps are displayed, see the Materials and Methods section. (a) Karyomaps for chromosome 11 in three embryos from a preimplantation genetic diagnosis (PGD) case for β-thalassemia and the unaffected child born following transfer of Embryo 1. The sibling used as a reference to phase the SNP calls is a carrier of the affected paternal allele (blue/orange), whereas Embryo 1 and the child born following the transfer of Embryo 1 are unaffected (red/orange), Embryo 2 is affected (blue/green), and Embryo 3 is a carrier of the paternal allele (blue/orange). Note the consistent pattern of key and non-key SNPs (colored dots) above and below the predicted haploblocks. In addition, note the crossovers in both the paternal (upper) and maternal (lower) chromosomes, common to all samples, indicating crossovers in the reference (boxed). (b) A detailed view of the β-globin locus (HBB) in Embryo 3, including the 2-Mb flanking regions proximal and distal to the gene (gray shading). Note that there are two isolated key SNPs (red dots), presumed miscalls, immediately distal to the gene. (c) Detailed karyomap of the huntingtin locus (HTT), with 2-Mb flanking regions to the left and right, in two single cells from a cleavage-stage embryo in a PGD case for Huntington disease. Note the recombination of the paternal chromosome (top) in the 2-Mb 3′-flanking region. The location of this recombination upstream of the gene is fixed by the presence of three non-key SNPs (in Cell 1) and by one key SNP and three non-key SNPs (in Cell 2), supporting the presence of the same haplotype as the reference. SNP, single-nucleotide polymorphism.