Figure 5.
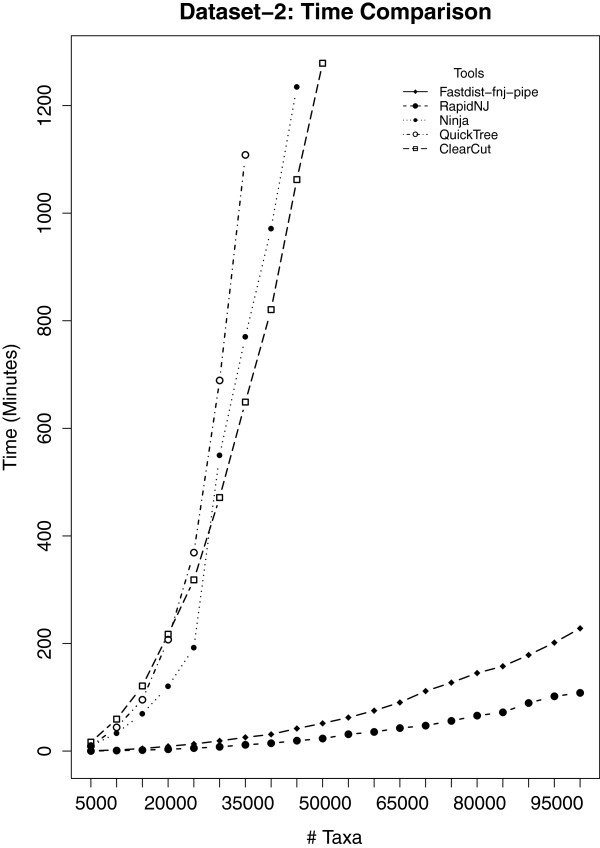
Time comparison analysis for dataset-2. Twenty gene families were considered for this analysis, with gene family members ranging from 5,000 to 100,000. The results suggest that fastphylo (fastdist-fnj-pipe) outperforms all the NJ-based tools considered in this study, except RapidNJ. QuickTree took the longest time to compute phylogenetic trees. In the allowed experimental time, i.e. 24 hours, QuickTree was able to compute phylogenetic trees for 7 gene families of size ranging from 5,000 to 35,500 family members, Ninja did it for 9 gene families with family members limited to 45,500, and ClearCut computed trees for 10 gene families (family size limited to 50,000). RapidNJ and fastphylo (fastdist-fnj-pipe), however, computed phylogenetic trees for all the 20 gene families considered in this study, in a reasonable time.
