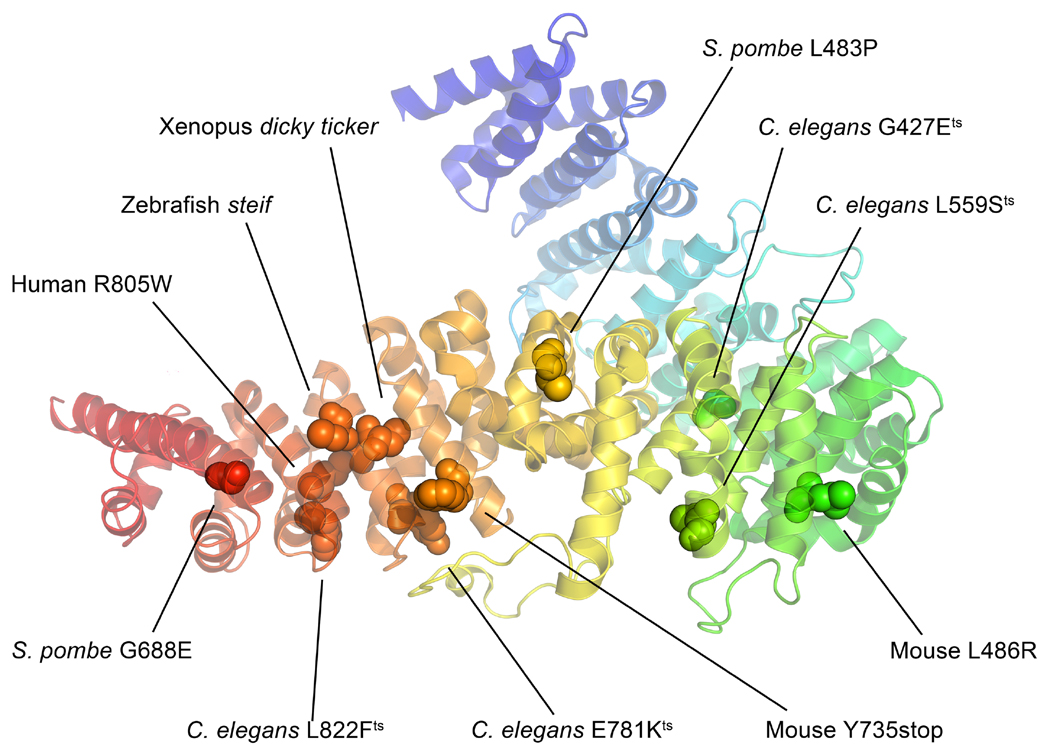
Figure 7. Mutations from the various UCS domain containing proteins mapped onto the Drosophila UNC-45 structure (PDBID 3NOW).
Mutations from S. pombe, C. elegans, zebrafish, Xenopus, mouse and human were mapped onto the Drosophila UNC-45 crystal structure using the protein alignment in Figure 6. A majority of the mutations is located in the UCS domain at the bottom of the figure, which underscores the importance of this domain to UNC-45 function. Residue numbers correspond to those of the mutant organisms. Protein structure modeled in PyMOL (PyMOL Molecular Graphics System, Version 1.6.0.0 Schrödinger LLC).