Use of herbicide-resistant rice cultivars in the United States has led to the emergence of herbicide-resistant weedy rice formed predominantly by hybridization of cultivars with historical weeds characterized by black hulls and awns.
Abstract
The use of herbicide-resistant (HR) Clearfield rice (Oryza sativa) to control weedy rice has increased in the past 12 years to constitute about 60% of rice acreage in Arkansas, where most U.S. rice is grown. To assess the impact of HR cultivated rice on the herbicide resistance and population structure of weedy rice, weedy samples were collected from commercial fields with a history of Clearfield rice. Panicles from each weedy type were harvested and tested for resistance to imazethapyr. The majority of plants sampled had at least 20% resistant offspring. These resistant weeds were 97 to 199 cm tall and initiated flowering from 78 to 128 d, generally later than recorded for accessions collected prior to the widespread use of Clearfield rice (i.e. historical accessions). Whereas the majority (70%) of historical accessions had straw-colored hulls, only 30% of contemporary HR weedy rice had straw-colored hulls. Analysis of genotyping-by-sequencing data showed that HR weeds were not genetically structured according to hull color, whereas historical weedy rice was separated into straw-hull and black-hull populations. A significant portion of the local rice crop genome was introgressed into HR weedy rice, which was rare in historical weedy accessions. Admixture analyses showed that HR weeds tend to possess crop haplotypes in the portion of chromosome 2 containing the ACETOLACTATE SYNTHASE gene, which confers herbicide resistance to Clearfield rice. Thus, U.S. HR weedy rice is a distinct population relative to historical weedy rice and shows modifications in morphology and phenology that are relevant to weed management.
Weedy rice (Oryza sativa), a conspecific weed of cultivated rice, is a global threat to rice production (Delouche et al., 2007). Classified as the same species as cultivated rice, it is highly competitive (Diarra et al., 1985; Pantone and Baker, 1991; Burgos et al., 2006), difficult to control without damaging cultivated rice, and can cause almost total crop failure (Diarra et al., 1985). The competition of cultivated rice with weedy rice can lead to yield losses from less than 5% to 100% (Kwon et al., 1991; Watanabe et al., 2000; Chen et al., 2004; Ottis et al., 2005; Shivrain et al., 2009b). Besides being difficult to control, weedy rice persists in rice fields because of key weedy traits, including variable emergence (Shivrain et al., 2009b), high degree of seed shattering (Eleftherohorinos, et al., 2002; Thurber et al., 2010), high diversity in seed dormancy (Do Lago, 1982; Noldin, 1995; Vidotto and Ferrero, 2000; Burgos et al., 2011; Tseng et al., 2013), and its seed longevity in soil (Goss and Brown, 1939). Weedy rice is a problem mainly in regions with large farm sizes where direct-seeded rice culture is practiced (Delouche et al., 2007). It is not a major problem in transplanted rice culture, where roguing weeds is possible and hand labor is available. The severity of the problem has increased in recent decades because of the significant shift to direct seeding from transplanting (Pandey and Velasco, 2002; Rao et al., 2007; Chauhan et al., 2013), which is driven by water scarcity (Kummu et al., 2010; Turral et al., 2011), increasing labor costs, and migration of labor to urban areas (Grimm et al., 2008).
The herbicide-resistant (HR) Clearfield rice technology (Croughan, 2003) provides an option to control weedy rice in rice using imidazolinone herbicides, in particular, imazethapyr. Imidazolinones belong to group 2 herbicides, also known as ACETOLACTATE SYNTHASE (ALS) inhibitors. Examples of herbicides in this group are imazamox, imazapic, imazaquin, and imazethapyr. Developed through mutagenesis of the ALS locus (Croughan, 1998), Clearfield rice was first commercialized in 2002 in the southern U.S. rice belt (Tan et al., 2005). Low levels of natural hybridization are known to occur between the crop and weedy rice. Gene flow generally ranges from 0.003% to 0.25% (Noldin et al., 2002; Song et al., 2003; Messeguer et al., 2004; Gealy, 2005; Shivrain et al., 2007, 2008). After the adoption of Clearfield technology, resistant weedy outcrosses were soon detected in commercial fields (Fig. 1), generally after two cropping seasons of Clearfield rice, where escaped weedy rice was able to produce seed (Zhang et al., 2006; Burgos et al., 2007, 2008). Similar observations have been reported outside the United States, in other regions adopting the technology (Gressel and Valverde, 2009; Busconi et al., 2012).
Figure 1.
Suspected herbicide-resistant weedy rice in a rice field previously planted with Clearfield rice along the Mississippi River Delta in Arkansas. More than 10 morphotypes of weedy rice were observed in this field, with different maturity periods. In the foreground is a typical weedy rice with pale green leaves; the rice cultivar has dark green leaves. The inset shows a weedy morphotype that matured earlier than cultivated rice.
Despite this complication, the adoption of Clearfield rice technology is increasing, albeit at a slower pace than that of glyphosate-resistant crops. After a decade of commercialization, 57% of the rice area in Arkansas was planted with Clearfield rice cultivars in 2013 (J. Hardke, personal communication). Clearfield technology has been very successful at controlling weedy rice, and polls among rice growers suggest that farmers have kept the problem of HR weeds in check by following the recommended stewardship practices (Burgos et al., 2008). The most notable of these are (1) implementation of herbicide programs that incorporate all possible modes of action available for rice production; (2) ensuring maximum efficacy of the herbicides used; (3) preventing seed production from escaped weedy rice, remnant weedy rice after crop harvest, or volunteer rice and weedy rice in the next crop cycle; (4) rotating Clearfield rice with other crops to break the weedy rice cycle; and (5) practicing zero tillage to avoid burying HR weedy rice seed (Burgos et al., 2008).
Clearfield rice has gained a foothold in Asia, where rice cultivation originated (Londo and Schaal, 2007; Zong et al., 2007). Clearfield rice received government support for commercialization in Malaysia in 2010 (Azmi et al., 2012) because of the severity of the weedy rice problem there. Dramatic increases in rice yields (from 3.5 to 7 metric tons ha−1) were reported in Malaysia where Clearfield rice was planted (Sudianto et al., 2013). However, the risk of gene flow and evolution of resistant weedy rice populations is high in the tropics, where up to three rice crops are planted each year, and freezing temperatures, which would reduce the density of volunteer plants, do not occur.
In the United States, where Clearfield technology originated and has been used for the longest time, the interaction between HR cultivated rice and weedy rice is not yet fully understood. Two main populations of weedy rice are known to occur in the southern United States and can be found in the same cultivated rice fields. These populations are genetically differentiated, are largely distinct at the phenotypic level, and have separate evolutionary origins (Reagon et al., 2010). One group tends to have straw-colored hulls and is referred to as the SH population; a second group tends to have black-colored hulls and awns and is referred to as the BHA population (Reagon et al., 2010). Genomic evidence suggests that both groups descended from cultivated ancestors but not from the tropical japonica subgroup varieties that are grown commercially in the United States. Instead, the SH group evolved from indica, a subgroup of rice commonly grown in the lowland tropics, and the BHA group descended from aus, a related cultivated subgroup typically grown in Bangladesh and the West Bengal region (Reagon et al., 2010). Weed-weed and weed-crop hybrids are also known to occur, but prior to Clearfield commercialization, these hybrids had occurred at low frequency (Reagon et al., 2010; Gealy et al., 2012). With the advent and increased adoption of Clearfield cultivars, the impact on U.S. weedy rice population structure and the prevalence of the SH and BHA groups are unknown.
Efforts to predict the possible consequences of HR or genetically modified rice on weedy rice have been a subject of discussion for many years. Both weedy rice and cultivated rice are primarily self-fertilizing, but, as mentioned above, low levels of gene flow are known to occur. Additional environmental and intrinsic genetic factors can act as prezygotic and postzygotic mating barriers between cultivated and weedy rice and influence the possibility and levels of gene flow between these groups (Craig et al., 2014; Thurber et al., 2014). However, once gene flow occurs between cultivated and weedy rice, and if the resulting hybrids are favored by selection, the resulting morphological, genetic, and physiological changes in weedy rice populations can alter the way that weedy rice evolves and competes. For example, herbicide-resistant weed outcrosses in an experimental field have been observed to be morphologically diverse (Shivrain et al., 2006), with some individuals carrying major weedy traits and well adapted to rice agriculture. Such weedy plants could be more problematic than their normal weedy counterparts. Thus, introgression of crop genes into weedy populations has the potential to change the population dynamic, genetic structure, and morphological profile of weedy plants. This, in turn, must alter our crop management practices. To increase our understanding of the impact of HR rice on the evolution of weedy rice, in this article we aim to (1) assess the frequency of herbicide resistance in weedy rice in southern U.S. rice fields with a history of Clearfield use; (2) characterize the weedy attributes of resistant populations; and (3) determine the genetic origins of herbicide-resistant weeds in U.S. fields.
RESULTS
Herbicide Resistance among Offspring of Weedy Rice in Clearfield Rice Fields
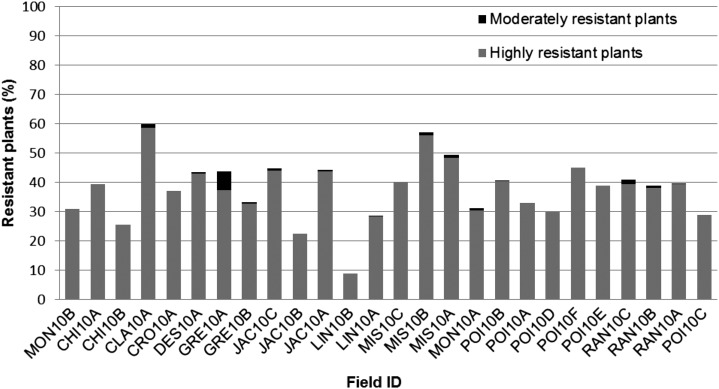
Twenty-six fields with a history of Clearfield rice use in Arkansas were sampled for weedy rice in the summer of 2010 (Fig. 1; Supplemental Table S1). Where weedy rice plants were detected at crop maturity, the density of weedy plants varied widely between 1 and 10 plants 1,000 m−2 (N.R. Burgos, unpublished data). These plants had survived two applications of imazethapyr as commercially practiced, early in the season, but not all were necessarily resistant. Among the 26 fields sampled, the collected weedy rice produced about 10% to 60% resistant offspring (Fig. 2). Nearly 100% of resistant plants were highly resistant to the commercial dose of imazethapyr, with no visible injury. Although resistant offspring were detected in collections from all fields sampled, averaged across fields, the majority of weedy rice offspring (63%) were still sensitive to imazethapyr.
Figure 2.
Frequency of weedy rice offspring resistant to the commercial dose of imazethapyr herbicide (two applications at 70 g ha−1) from weedy plants collected in fields previously planted with Clearfield rice. Moderately resistant plants present 21% to 79% injury, and highly resistant plants present 0% to 20% injury.
Phenotypic Characterization of Herbicide-Resistant Weedy Rice
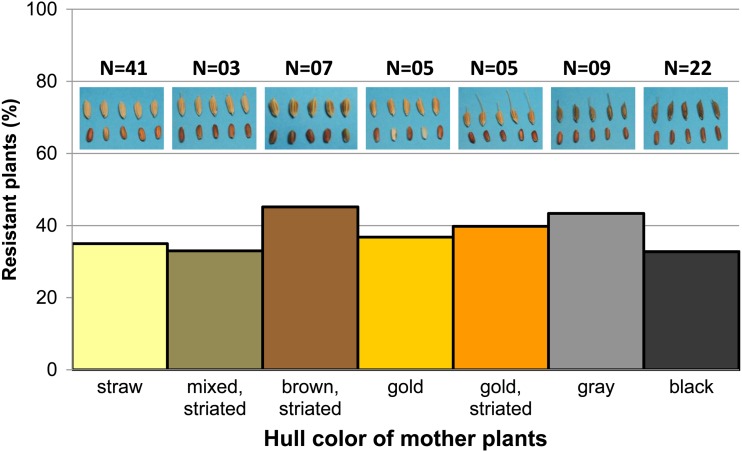
To determine if weeds that survive herbicide application are different from those observed historically in the United States prior to the widespread use of Clearfield rice (i.e. historical weeds), we phenotyped various traits in 720 offspring of 48 weedy rice accessions that we considered to be HR (Supplemental Table S2) and compared them with historical weeds. Among the traits commonly used to distinguish historical weedy populations is hull color. Weedy rice that escaped control measures in fields previously planted with Clearfield rice had the usual hull colors of black, brown, and straw but also had myriad color variations: gold, gray, and purple with or without striations (Fig. 3). This trait per se may not contribute to weediness but is indicative of the increased phenotypic diversification of weedy populations in fields with HR rice.
Figure 3.
Frequency of imazethapyr-resistant offspring of weedy rice harvested in 2010 from commercial fields with a history of Clearfield rice classified by the hull color of the mother plant.
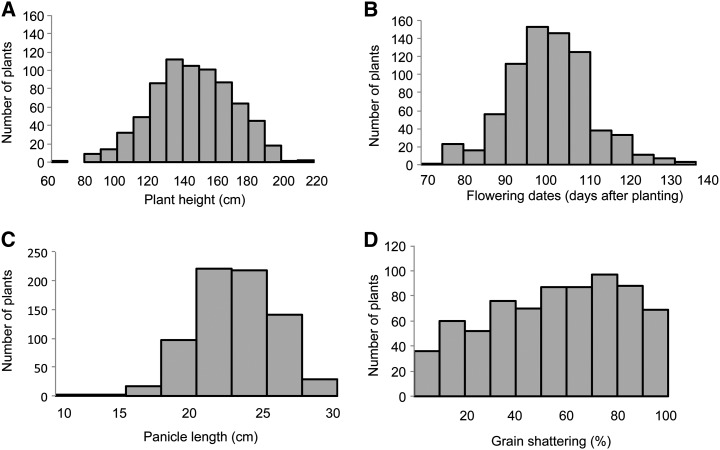
The majority of HR offspring of weedy rice exerted panicles between 90 and 110 d after planting (DAP; Fig. 4A), with an average heading date of 100 DAP. The resistant weedy rice were generally tall (Table I), with the majority being more than 1 m in height (Fig. 4B). A few were shorter than cultivated rice, as was observed with pre-Clearfield rice accessions (Gealy et al., 2006; Shivrain et al., 2010b). Panicles were mostly 20 to 25 cm long (Fig. 4C), similar to cultivated rice. Grain shattering ranged from minimal to 100% (Fig. 4D). Considering the weedy traits that strongly affect the phenotypic grouping of populations (heading date, plant height, flag leaf length, panicle length, awn length, and seed shattering), CLUSTER analysis was conducted and showed that the HR weedy rice plants separated into seven clusters (Table I). The largest group was cluster 1; it had intermediate height (141 cm) and early heading (93 DAP) relative to other groups, except cluster 3, and shattered relatively less (40%). The shortest plants were in clusters 5 and 6 (mean = 123–125 cm), with the same average heading date (105 DAP) but distinguished by high seed shattering in cluster 6 (76%) and low seed shattering in cluster 5 (34%).
Figure 4.
Frequency distribution of weedy rice traits resistant to the full dose of imazethapyr (70 g ha−1, two applications) at the Arkansas Rice Research and Extension Center.
Table I. Hierarchical clustering of herbicide-resistant weedy rice from Arkansas based on selected morphological traits and heading period.
| Cluster | No. | Headinga |
Flag Leaf Length |
Plant Heightb |
Panicle Lengthc |
Awn Lengthd |
Shattering |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | ||
| DAP | cm | % | |||||||||||
| 1 | 233 | 93 | 74–117 | 33 | 19–55 | 141 | 84–190 | 23 | 16–28 | 1.2 | 0–6.1 | 40 | 1–91 |
| 2 | 104 | 106 | 88–131 | 35 | 22–55 | 148 | 95–184 | 24 | 19–28 | 1.1 | 0–4.4 | 77 | 38–100 |
| 3 | 56 | 94 | 78–106 | 32 | 21–49 | 163 | 110–199 | 22 | 17–26 | 0.6 | 0–3.2 | 68 | 10–98 |
| 4 | 54 | 98 | 87–110 | 34 | 23–54 | 171 | 125–200 | 23 | 20–27 | 3.7 | 1.7–6.0 | 75 | 36–97 |
| 5 | 53 | 105 | 90–128 | 32 | 17–46 | 123 | 91–175 | 22 | 18–28 | 2.0 | 0–4.8 | 34 | 2–86 |
| 6 | 107 | 105 | 83–130 | 30 | 17–48 | 125 | 62–159 | 20 | 10–24 | 1.7 | 0–6.7 | 76 | 14–100 |
| 7 | 113 | 102 | 78–124 | 36 | 20–54 | 160 | 127–215 | 26 | 22–28 | 4.4 | 1.0–8.4 | 39 | 0–96 |
| lsd0.05 | 2 | 2 | 5 | 1 | 0.4 | 5 | |||||||
Recorded as DAP, when four tillers from a plant had exerted panicles. Up to five plants per plot were marked for characterization, based on the apparent morphological variation. bMeasured from the base of the panicle to the tip of the flag leaf of the main culm. cFive panicles were measured per plant. dDetermined by applying equal force to each sample, as described in “Materials and Methods.”
Seed shattering is a weedy trait that contributes greatly to weed persistence. The HR weedy plants grouped into four phenotypic clusters with respect to seed shattering (Table II). Group 1 (25% of plants) had the highest shattering capability (87%). These types of plants will drop almost all of their seeds prior to crop harvest and are expected to quickly dominate the soil seed bank if allowed to reproduce. Plants in group 2 (65% shattering) present a similar problem. Most of the seeds of plants in group 4 (15% shattering) are likely to be harvested and removed from the field with the crop; thus, this group is likely to make only a minor contribution to the soil seed bank. Such plants could be a problem in the short term because their seeds will contaminate the harvested rice grain and may result in dockage of the grain price if present in quantities above the allowable red rice contamination level (Ottis et al., 2005). The Clearfield rice cultivars we characterized in this test had 25% to 34% shattering (Table III), which was higher than some of the weedy outcrosses.
Table II. Hierarchical clustering of herbicide-resistant weedy rice based on seed shattering.
Shattering was measured as described in “Materials and Methods.”
| Cluster | No. of Accessions | Proportion of Total Accessions | Mean Shattering | Range |
|---|---|---|---|---|
| % | ||||
| 1 | 41 | 17 | 86 | 79–97 |
| 2 | 56 | 23 | 26 | 18–34 |
| 3 | 76 | 32 | 48 | 36–57 |
| 4 | 68 | 28 | 67 | 58–77 |
Table III. Grain shattering of herbicide-resistant Clearfield rice characterized along with the herbicide-resistant weedy rice.
Shattering was measured as described in “Materials and Methods.”
| Cultivar | Rice Type | No. | Proportion of Total Plants | Mean | sd | Range |
|---|---|---|---|---|---|---|
| % | ||||||
| CL151 | Inbred | 6 | 27 | 33 | 15 | 10–50 |
| CL XL729 | Hybrid | 9 | 41 | 25 | 21 | 1–52 |
| CL XL745 | Hybrid | 7 | 32 | 34 | 18 | 8–54 |
Seed dormancy exacerbates the persistence of shattered seeds. The HR weedy rice separated into three groups with respect to germination capacity (GC; Table IV). The GC varied widely among the resistant, weedy outcrosses, with averages of 20% to 100% germination at 75 and 270 d after harvest. A large majority of the HR weedy rice showed loss of seed dormancy, with a mean GC of 97% at 270 d after harvest. This was numerically higher than the mean GC of rice cultivars (88%–94%) at the same period (Table V). Group 2 of HR weedy rice had lower mean GC (85%) than group 1 (Table IV), but this dormancy is low relative to what is generally observed in historical weedy populations (Tseng et al., 2013). Only a small proportion of HR weedy rice, group 3 (10% of 205 plants characterized), showed significant dormancy with about 50% GC after 270 d of afterripening.
Table IV. GC of herbicide-resistant weedy rice 75 and 270 d after harvest.
Seeds were afterripened at room temperature for the prescribed period and germinated in the dark at 30°C for 12 d.
| Group | No.a | Proportion of Total Plants | 75 d after Harvest |
270 d after Harvest |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | sd | Range | Mean | sd | Range | |||
| % | ||||||||
| 1 | 99 | 16 | 65 | 13 | 24–88 | 85 | 10 | 66–100 |
| 2 | 463 | 74 | 94 | 5 | 76–100 | 97 | 4 | 74–100 |
| 3 | 63 | 10 | 76 | 18 | 20–100 | 51 | 13 | 20–69 |
No. represents three replicates of 89 weedy rice accessions, with up to five plants characterized per accession in a field experiment at Stuttgart, AR, in three replicates.
Table V. GC of herbicide-resistant cultivated rice 75 and 270 d after harvest.
Seeds were afterripened at room temperature for the prescribed period and germinated in the dark at 30°C for 12 d.
| Cultivar | No. | 75 d after Harvest |
270 d after Harvest |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | sd | Range | Mean | sd | Range | ||||
| % | |||||||||
| CL151 | 6 | 89 | 9 | 79–100 | 91 | 16 | 67–100 | ||
| CL XL729 | 9 | 85 | 9 | 70–100 | 88 | 11 | 70–100 | ||
| CL XL745 | 7 | 95 | 6 | 83–100 | 94 | 6 | 83–100 | ||
Population Structure of Herbicide-Resistant Weedy Rice
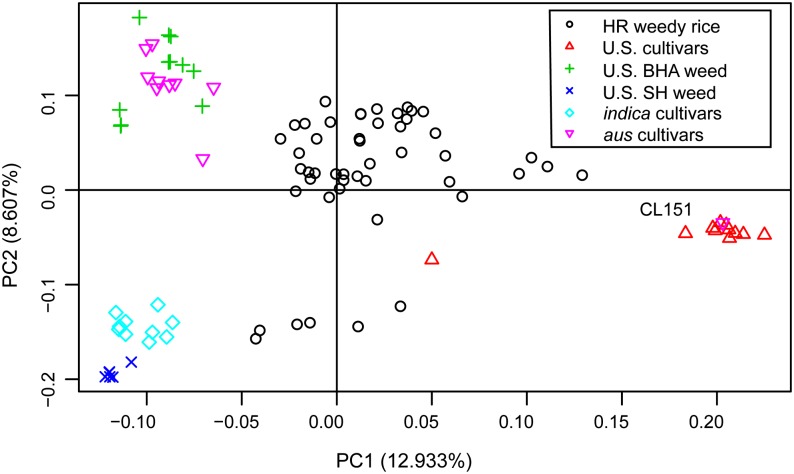
To determine to what extent HR offspring of weeds collected in Clearfield fields resemble U.S. weeds occurring prior to widespread Clearfield use (i.e. historical weeds) at the genetic level, 48 weedy accessions deemed HR in our phenotyping trials were selected for genotyping (Supplemental Table S2). Using genotyping by sequencing (GBS; Elshire et al., 2011), we characterized single-nucleotide polymorphisms (SNPs) across the genome in samples of HR weedy rice, U.S. and Asian cultivated rice, historical weeds (Reagon et al., 2010), and wild species of Oryza (Supplemental Table S3). Principal component analysis, which excluded wild Oryza spp. samples, showed that the HR weedy rice, regardless of hull color, clusters separately from other weedy rice in the United States, from Asian cultivars, and from U.S. cultivars (Fig. 5). The historical U.S. BHA weedy rice clusters with aus rice from South Asia, consistent with its known ancestral origins (Reagon et al., 2010). Similarly, historical U.S. SH weedy rice forms a cluster that is closely related to indica cultivars from Asia. U.S. rice cultivars form a distinct, tight cluster indicative of high genetic similarity among U.S. germplasm. The herbicide-resistant reference, cv CL151, clusters with U.S. cultivars, as expected, because it was developed with U.S. germplasm. An HR long-grain cultivated hybrid rice lies between U.S. cultivars and the indica-SH cluster (Fig. 5), which is consistent with the findings for a long-grain hybrid in an earlier report (Gealy et al., 2009).
Figure 5.
Principal component analysis of HR weedy rice, U.S. cultivated rice, historical SH and BHA weedy rice, and Asian aus and indica cultivars. Principal component 1 (PC1) explains 12.93% of the variance, and PC2 explains 8.61%. The inbred reference Clearfield cultivar, CL151, is labeled.
Although contemporary HR weeds form a distinct cluster, this clustering is situated between U.S. cultivars and weed/Asian cultivars and is somewhat diffuse (Fig. 5). The intermediate location of HR weeds is suggestive of hybridization between historical U.S. weeds and U.S. cultivars. The large size of the cluster indicates high genetic diversity among these plants relative to the other groups and is consistent with the observed phenotypic and phenological diversity of HR weedy rice.
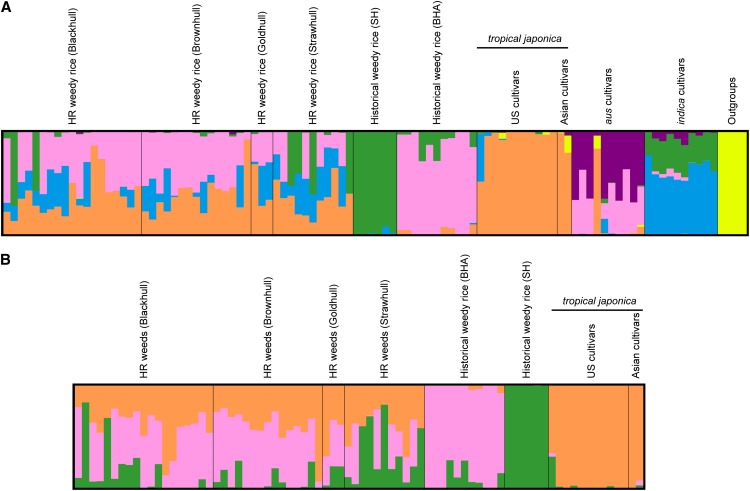
To further investigate the population structure of historical weedy rice and HR weeds in the United States, we carried out hierarchical STRUCTURE (Pritchard et al., 2000) analyses. A first analysis included the same sampling of rice groups as above as well as wild rice samples (Supplemental Table S3). The Evanno method (Evanno et al., 2005) identified the best model as one containing two populations (K; Supplemental Table S4). Because the Evanno method can often underestimate K when there is hierarchical population structure (Waples and Gaggiotti, 2006), and because numerous studies have found population differentiation between the historical weedy groups, between indica and aus varieties, and between wild species of Oryza and rice (Garris et al., 2005; Caicedo et al., 2007; Reagon et al., 2010), we also examined likelihood scores. The highest likelihood score for the number of clusters (K) was six (Fig. 6A; Supplemental Table S4); this model is biologically consistent with previous characterizations of weedy and cultivated rice. In this model, the wild species of rice are highly distinct. The historical weedy SH and BHA from the United States are also distinct separate populations, supporting the principal component analysis result and as expected from previous characterizations (Reagon et al., 2010). The cultivated ancestors of historical U.S. weeds, the aus and indica subgroups, appear as distinct populations; however, these ancestors share some genetic identity with their descendant weed groups, as expected from their ancestral status. Japonica rice, which includes U.S. cultivars, is highly distinct from the other cultivated populations and those of historical weedy rice.
Figure 6.
Population structure of herbicide-resistant weedy rice and related groups based on GBS data. A, Population structure of HR weedy rice, weedy rice occurring prior to the widespread use of Clearfield rice (i.e. historical weedy rice), reference cultivars from Asia and the United States, and wild Oryza spp. B, Population structure of HR weedy rice, historical weedy populations, and tropical japonica cultivars.
In contrast, the contemporary HR weedy rice is clearly an admixed population. Contributions from japonica, historical BHA weedy rice, and either indica or historical SH weedy rice are clearly seen in most of the HR weedy individuals examined (Fig. 6A). Although the weedy contribution varies per individual, all HR weeds have a measure of japonica contribution and the great majority have a BHA contribution. Although smaller, a certain amount of either SH or indica contribution is also evident in many of the HR weeds (Fig. 6A).
We attempted a more focused characterization of HR weeds running STRUCTURE with only U.S. cultivars, historical weeds, and HR weeds, as these first groups are the ones most likely to have contributed directly to contemporary HR weeds. We focused on the K = 3 model, which can distinguish tropical japonica, SH, and BHA populations (Fig. 6B; Supplemental Table S5). In this reduced model, the HR weedy rice again is clearly an admixed population of crops and historical weeds. The disproportionate contribution of historical BHA weeds to HR weedy rice is also confirmed: all but six HR weedy rice carry a measurable proportion of the BHA genome (Fig. 6B).
Correlations between Population Structure and Phenotype in HR Weedy Rice
Whereas hull color was among the morphological traits most strongly correlated with population structure in historical U.S. weedy rice (Reagon et al., 2010; Shivrain et al., 2010a), no clear correlation between hull color and the population structure of HR weedy rice was observed (Fig. 6). However, the results in Figure 6B suggest that there was less introgression of SH alleles into HR black-hull and brown-hull individuals and that most HR individuals with low BHA contribution tend to have straw-colored hulls. Population structure by genotype also did not correlate strongly with any other morphological groupings in HR weeds (Tables I, II, and IV).
Parental Contributions to the Origin of HR Weedy Rice
To assess the parental contributions to U.S. HR contemporary weedy rice, we examined locus-specific population differentiation between HR weeds and historical weeds in SNPs genotyped across the genome using the program LOSITAN (Beaumont and Nichols, 1996; Antao et al., 2008). The average fixation index (FST) between contemporary and historical weeds is 0.219, indicating moderate differentiation. The top 95 FST values observed in the data set, however, ranged from 0.759 to 0.895 and were all found for SNPs located on chromosome 2, in a region ranging from approximately 11 to 19 Mb (Supplemental Table S6). Of the 4,764 loci with FST values in the top confidence interval (P > 0.975), 794 fall on chromosome 2, the most of any chromosome. The ALS gene that confers resistance to Clearfield cultivated rice varieties lies approximately 18 Mb on chromosome 2 (Sales et al., 2008). This suggests that contemporary HR weedy rice possess the cultivated allele of ALS, leading to herbicide resistance and high differentiation from ancestral weeds in this region of chromosome 2.
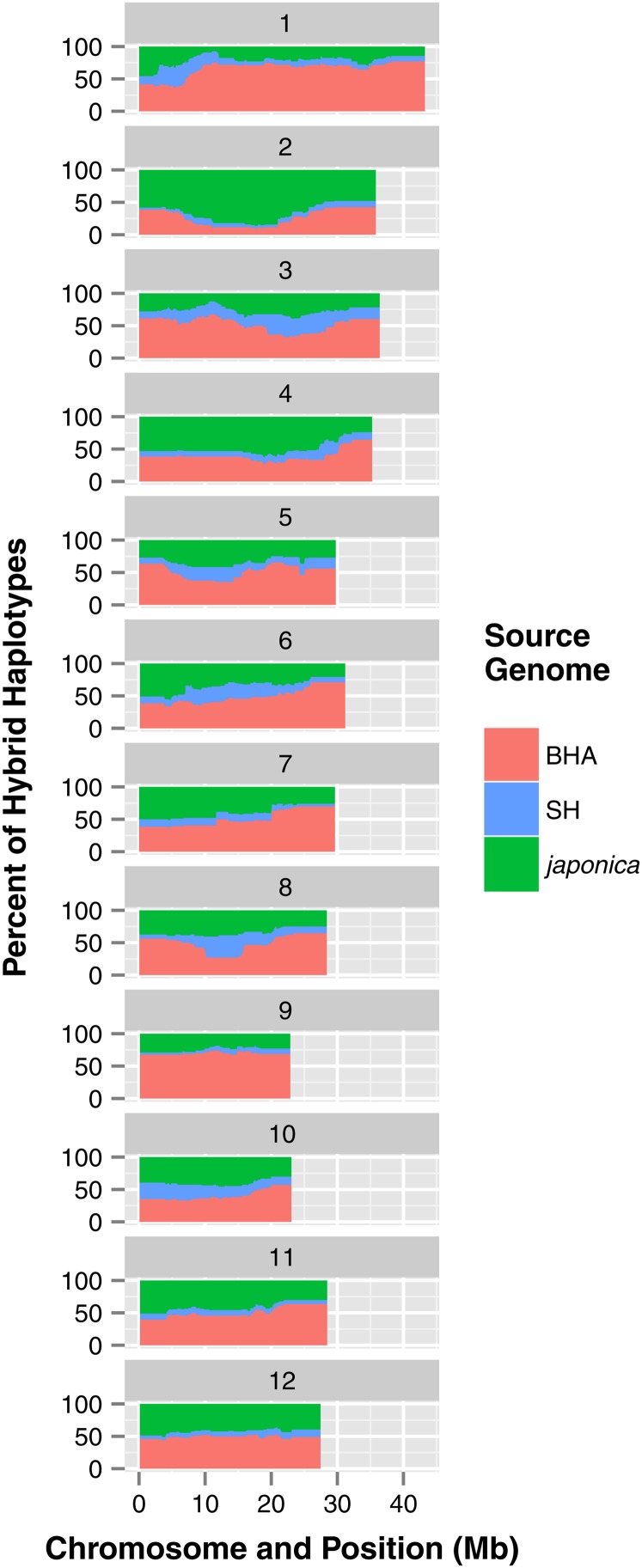
To better characterize the portions of the genome that have been contributed by each ancestral gene pool to the new population of admixed HR weeds, we used LAMP-LD, a program that takes into account linkage disequilibrium information in ancestral reference populations to infer the ancestry of local areas of the genome. We stipulated the historical SH, historical BHA, and japonica samples as mixing populations. We then searched for contiguous runs of loci (SNPs) for which 80% or more of the contemporary weed individuals have at least one japonica allele. Very few regions in the genome of HR weeds were dominated by japonica or any other ancestral allele, indicating primarily random mixing of the ancestries in admixed individuals. The exceptions were from positions 6,748,749 to 22,174,942 bp on chromosome 2 and from positions 20,448,681 to 20,855,557 bp on chromosome 4; both of these regions were overrepresented for japonica alleles in the HR population (Fig. 7; Supplemental Table S7). The first region on chromosome 2 encompasses the known location of the ALS gene (LOC_Os02g30630), and japonica Clearfield ALS alleles are likely to provide the resistance to herbicide observed in this contemporary weedy rice population. We do not know of a gene necessary for herbicide resistance on chromosome 4, but it could be that this narrower genomic region contains one or more genes where crop alleles could be favorable for weed adaptation.
Figure 7.
Distribution of parental population contribution to the HR weedy rice population as determined by LAMP-LD. Each horizontal bar represents a single chromosome. Colors denote possible parental populations. For all SNPs included in the analysis, the percentage of haplotypes carrying alleles from each parental population is shown.
We also detected some genomic regions where crop alleles were underrepresented and weed alleles were thus overrepresented. For example, three regions on chromosome 1 (27,481,759–27,690,136 bp; 29,397,914–32,089,170 bp; and 34,787,952–43,243,943 bp) have contiguous runs of SNPs where 80% or more HR weeds carry BHA alleles (Fig. 7; Supplemental Table S7). Other areas in the genome that stand out due to the overrepresentation of individuals with historical weed alleles are 14,886,856 to 15,653,975 bp and 17,509,003 to 17,658,793 bp on chromosome 9. These regions may possess genes where weed alleles are necessary or advantageous for weediness.
DISCUSSION
Weedy rice has long been one of the most damaging weeds in direct-seeded rice cropping systems, such as that employed in the United States. Because it is the same species as cultivated rice, it has historically been very difficult to control weedy rice without also damaging the crop. In the last decade, development of the herbicide-resistant Clearfield rice technology (Croughan, 2003) has provided an option to control weedy rice in rice using imidazolinone herbicides. Since the adoption of Clearfield technology, herbicide-resistant weedy rice plants have been detected in U.S. rice fields (Zhang et al., 2006; Burgos et al., 2007, 2008), but the impact of Clearfield use on weedy rice at the population level has not been examined. Here, we attempted to assess the impact of Clearfield technology on the evolution of weedy rice, an essential aspect to the design of appropriate weed-control strategies.
The weedy rice infestations in the 26 fields with Clearfield rice history we sampled were generally what rice growers and consultants consider light (Burgos et al., 2008), indicating a reduction in weedy rice infestation. Growers normally plant Clearfield rice in fields heavily infested with weedy rice. Between 10% and 60% of offspring of weedy rice from these fields were found to be HR (Fig. 2). It is important to note that the frequency of resistant individuals we detected was artificially high, because we targeted fields that contained remnant weedy rice after having been sprayed with imazethapyr. The only susceptible plants that would have remained in these fields would be those that happened to escape the herbicide application by emerging late or by having received a sublethal dose, as could happen along field edges. The resistance frequency would have been drastically lower if germinating seedlings were sampled at the beginning of the season, prior to imazethapyr application, instead of sampling mature plants at the end of the season. However, our objectives were to assess how much the surviving weedy rice plants could potentially contribute to resistance in future weed populations and what differences could be detected between these remnant weedy plants and weeds occurring historically in the United States. Our results show that a substantial proportion of seeds produced by surviving weedy rice in Clearfield fields is HR.
From seed produced by surviving weedy rice, we selected only HR plants to characterize in morphology, phenology, and genetics. These plants are thus not representative of all weedy rice occurring currently in the United States or even all weedy rice found in Clearfield rice fields. These HR weeds, however, are representative of the type of weed that is most likely to be favored with continued use of imidazolinone herbicides. Continued or excessive use of Clearfield technology could lead to the prevalence of weedy populations descendant from HR weedy accessions such as the ones characterized in this study.
Our study shows that contemporary HR weeds are clearly an admixture between historical weeds and U.S. cultivars, the latter probably being Clearfield rice (Figs. 5 and 6). Our results suggest, moreover, that with selection of HR weedy rice, phenotypic and genetic diversity may increase in weedy populations. For example, in the past, weedy rice in the United States has comprised primarily awnless straw-hull types (70%), with black-hull types comprising only 22% and brown-hull types comprising only 7% of historical weedy populations (Shivrain et al., 2010b). We observed more seed types from HR weedy rice populations (Fig. 3) than among the historical populations (Shivrain et al., 2010b). Likewise, heights of HR weedy rice were widely diverse, from 0.6 to 2 m tall, while historically weedy rice plants have been generally tall (mean = 125 cm, 200+ accessions), with the tallest plants being 190 cm (Shivrain et al., 2010b). This height diversity is important for management because short plants escape notice by field scouts. Tall plants, on the other hand, are voracious consumers of fertilizer and formidable competitors with rice (Burgos et al., 2006); tall plants also shade the crop and will lodge, bringing down adjacent rice plants and causing harvest losses.
Not all of the admixed HR weeds we characterized necessarily have ideal weedy traits. For example, some HR plants exhibit complete or nearly complete loss of shattering (Table II), which is not typical of historical weeds (Thurber et al., 2010). A high degree of shattering is advantageous to weeds because it allows replenishment of the soil seed bank before the crop is harvested, promoting persistence. In contrast, crops have been bred to minimize shattering to increase harvestable yield. Most seeds from low-shattering phenotypes, such as those of groups 2 and 3, which comprised almost 50% of the HR weedy population (Table II), are likely to be harvested with the crop. Although in the short term these could be detrimental to grain quality and would reduce profitability, these weeds are likely to be eventually culled from the population. However, it should be noted that a small amount of all harvested grain is inevitably lost back onto the field due to mechanical inefficiencies of the threshing process.
Despite the presence of low-shattering weeds, at least 50% of HR weeds exhibit high levels of shattering, with averages of at least 65%. Therefore, abundant HR seeds are being deposited into the soil seed bank. Moreover, even for HR weeds with low shattering, any plants that remain after the crop season ends can backcross repeatedly to other weedy plants if allowed to proliferate, which eventually will restore the shattering trait. This is possible because the majority of HR weedy rice have overlapping flowering periods (Table I) as well as a long duration of flowering time (4–17 d; Shivrain et al., 2009b). Collectively, the HR population flowered later than historical weeds, which average 57 to 99 DAP (Shivrain et al., 2010b). About 75% of HR weedy rice also flowered later than reference Clearfield cultivars (87 and 93 DAP, respectively, for the hybrid and inbred cultivars), but not late enough to prevent ample reseeding from the weed. An extremely late maturation period is detrimental to weedy rice because it reduces seed production potential. Low night temperatures (in late summer to fall), for example, reduce pollination and spikelet fertility, making seed set difficult (Shimono et al., 2005), and late-maturing plants may also be eliminated during or after harvest.
A highly important weedy trait is seed dormancy. Historical weedy rice populations exhibit great diversity in seed dormancy (Burgos et al., 2011; Tseng et al., 2013) but are predominantly dormant. In contrast, nearly 75% of the HR weedy rice plants were nondormant (Table IV). Although this may suggest that the majority of HR weedy rice genotypes we observed will not persist, it takes only a few individuals to initiate a highly adapted population, such as those in group 3 (Table IV). Moreover, outcrosses between weedy types could reintroduce the dormancy trait into some weedy genotypes.
Although both cultivated rice and weedy rice are primarily self-fertilizing (Beachell et al., 1938), it is clear from previous results and ours that low levels of gene flow can occur and, if favored by selection, can lead to the creation of new types of weeds. Although it is perhaps not surprising that crop-weed outcrosses have given rise to HR weeds in fields where HR cultivated rice has been used, one interesting aspect of the genetic background of HR weeds we sampled was the predominant contribution of BHA historical weeds. Of the 48 weed accessions characterized with GBS, only six seemed to not have any BHA contribution based on STRUCTURE results (Fig. 6), and portions of the BHA genome were overrepresented in the HR population, while no portion of the SH genome was (Fig. 7; Supplemental Table S5).
Interestingly, while the SH genetic contribution to HR weeds did not predominate, some level of contribution from either indica cultivars or SH weeds was evident in most weeds sampled (Fig. 6A). Indica cultivars are not grown commercially in the United States, so a direct contribution of indica to the genome of HR weedy rice is unlikely. The suggestion of indica ancestry in some HR weeds can likely be attributed to one of two explanations. It could be that our STRUCTURE analysis cannot accurately distinguish indica from the SH background due to their shared ancestry (Reagon et al., 2010). This would suggest that many HR weeds are really the product of three-way outcrosses involving cultivated rice, SH, and BHA. Alternatively, some of the outcrosses leading to HR weedy rice may have involved hybrid Clearfield lines; high-yielding hybrid Clearfield cultivars have been introduced in recent years and reportedly possess indica genomic contributions, which could be passed into the weed’s genetic background. If this latter explanation is true, only six of our HR weedy accessions would possess a true SH contribution (Fig. 6), again underscoring the smaller role this group seems to have played in the evolution of HR weedy rice. Moreover, these results would suggest that crop-weed outcrossing is more likely to occur with hybrid rice varieties, as shown by Shivrain et al. (2009c)
Various postzygotic and prezygotic barriers can influence the amount of gene flow that occurs between cultivated rice and weedy rice, and these may vary depending on the cultivar grown in a field. In the United States, weedy and cultivated rice belong to separate domesticated rice lineages (indica and japonica, respectively; Reagon et al., 2010), and various genetically determined postzygotic mating barriers are known to occur between individuals of these lineages (Ouyang and Zhang, 2013). However, a recent study examining hybrid sterility genes that could act as gene flow barriers between U.S. historical weeds and U.S. cultivars found many fewer barriers than expected due to ancestry (Craig et al., 2014). Although the probability of partial hybrid sterility was a bit higher for BHA weeds than SH weeds crossing with japonica cultivars, the distribution of hybrid sterility alleles in these groups suggested that hybrid sterility is unlikely to limit gene flow between the crop and all weed groups.
Controlled crosses done with cv CL161 (a tropical japonica inbred Clearfield cultivar) and 12 historical weedy rice ecotypes of varying hull characteristics (seven straw-hull, three black-hull, and two brown-hull ecotypes) did not show strong barriers to hybridization for any hull color (Shivrain et al., 2008). Seed set ranged from 70% to 94% except for two straw-hull ecotypes, which showed less genetic compatibility with the rice cultivar. In field experiments carried out in Stuttgart, Arkansas, when the same 12 ecotypes were allowed to overlap in flowering with cv CL161, the highest outcrossing rate (0.25%) was observed with one straw-hull ecotype, followed by two black-hull and two brown-hull ecotypes (Shivrain et al., 2008), supporting the ease with which BHA is likely to cross with cultivated rice. Another factor possibly contributing to higher outcrossing between BHA and cultivated rice is the low GC (high dormancy) of black-hull awned weedy rice (Tseng et al., 2013), as weedy plants that germinate almost 100% in one season could be controlled 100% in that season.
Studies of flowering time, a potentially powerful prezygotic mating barrier, in U.S. weedy and cultivated rice have also shown that greater overlap in flowering time exists between U.S. crops and BHA weeds than with SH weeds (Reagon et al., 2011; Thurber et al., 2014). Moreover, U.S. crops and BHA weeds share nonfunctional alleles of Heading Date1, the major gene controlling flowering time in rice, which render the majority of the individuals in these populations daylength insensitive (Thurber et al., 2014). These shared nonfunctional alleles make it more likely that BHA weeds and U.S. crops will flower simultaneously, which may lead to greater amounts of gene flow between these groups, and could account for the greater BHA contribution to HR weeds. Moreover, the flowering of first-generation hybrids produced from controlled crosses between straw-hull weedy rice (as pollen source or pollen receiver) and HR or non-HR rice cultivars can be delayed from 2 to 5 weeks compared with both parents (Gealy et al., 2006; Shivrain et al., 2009a), whereas hybrids produced from crossing rice with black-hull or other awned weedy rice have exhibited no such delay in flowering (Gealy et al., 2006). If late-season temperatures begin to fall appreciably, this delay can greatly reduce the production of viable seeds in crosses with straw-hull weeds, which may provide another explanation for the greater frequency of BHA hybrids observed in our study.
Given that the 48 HR weeds we characterized and genotyped were collected from 13 different fields, it is likely that many of these weeds represent separate origins and are the products of multiple outcrosses with cultivated rice. Multiple outcrosses, coupled with the independent assortment and recombination that would occur with further selfing, are likely to give rise to a variety of admixed genotypes. Given this, it is striking that a few genomic regions from a given parental population are overrepresented in HR weeds. The most striking of these regions covers approximately 15 Mb on chromosome 2, where the great majority of HR weeds carry japonica alleles (Fig. 7; Supplemental Tables S6 and S7). This region encompasses the known location of the ALS gene, suggesting that introgressed alleles of the Clearfield ALS gene are likely the source of herbicide resistance in HR weedy rice. Crop alleles were also overrepresented in HR weeds in approximately 600 kb on chromosome 4 (Fig. 7; Supplemental Table S7), and BHA alleles are overrepresented in several portions of chromosome 10, including approximately 9 Mb near the chromosome’s end, and some smaller genomic regions on chromosome 9. The current significance of these highly represented regions is not known, but it is possible that their pervasive presence in HR weeds is due to the importance of given crop or weed alleles in these genomic regions to weed adaptation.
CONCLUSION
The increased use of HR Clearfield rice cultivars in the last decade has changed the selective landscape for weedy rice. Fields with a history of Clearfield rice planting invariably have HR weedy rice, due to hybridization with the crop. Hybridization seems to be occurring primarily between BHA historical weeds and hybrid Clearfield varieties, but this does not preclude hybridization with other groups. Overall, hybridization with rice cultivars increases the diversity of weedy populations, although the frequency of resistant plants is relatively low. Some encouraging facts emerged from this study. A large proportion of HR weedy rice exhibit reduced weediness in terms of reduced seed dormancy, later flowering (and hence more probability of reduced seed production), and reduced seed shattering, which reduces persistence. If the recommended best management practices are followed, it is possible that the proliferation of HR weedy rice can be limited. However, the continued occurrence of outcrosses between weedy rice and HR cultivars is a risk for the emergence of more competitive weedy rice types.
MATERIALS AND METHODS
Plant Materials and Experimental Design
In the summer of 2010, we collected weedy rice (Oryza sativa) panicles from 26 fields with a history of Clearfield rice in Arkansas, spanning 15 counties (Supplemental Fig. S1; Supplemental Table S1). Up to 20 panicles (two per plant) were collected for each plant morphotype present in the field and bulked. When only a few plants represented a plant morphotype, all panicles from these plants were collected. Each type collected from the farmers’ fields is referred to as an accession. The panicles were hand threshed, and the grain phenotype (hull color and awn length) was recorded. Seeds were stored for 9 months and planted at the Arkansas Rice Research and Extension Center (Stuttgart) in 2011 to test for resistance to imazethapyr herbicide. Due to the length of storage, the great majority of seeds had already broken primary dormancy (Tseng et al., 2013). Eighty-nine weedy rice accessions were tested. The experiment was conducted in a split-plot arrangement with herbicide treatment (nontreated, half dose, and full dose) as the main plot and weedy rice accession as subplot, with three replications. Fifty seeds from each accession were drill seeded in single rows, 4.6 m long and 0.6 m apart, on May 20. Nitrogen (28 kg ha−1) was applied at planting and immediately before permanent flooding. For general weed control, quinclorac (0.42 kg ha−1) and pendimethalin (1.12 kg ha−1) were applied at 5 DAP. For full-dose herbicide treatment, imazethapyr (0.07 kg ha−1) plus 0.24% (v/v) nonionic surfactant was applied in a volume of 150 L ha−1 using a tractor sprayer following commercial recommendation, first when rice was at the V3 stage (Counce et al., 2000) and again 7 d later.
Resistance Evaluation and Phenotypic Characterization of Resistant Plants
Plant response (injury and mortality) was evaluated 21 d after the second herbicide application. This was used to determine the frequency of resistant plants (Fig. 1). To calculate percentage mortality, the number of plants per row was counted before herbicide application and used as a reference for the number of dead plants 21 d after herbicide application. Injury incurred by the surviving plants was assessed visually on a scale of 0% (no visible injury) to 99% (barely alive). Surviving plants with at least 80% injury were classified as susceptible; otherwise, they were considered resistant (Supplemental Table S1).
Up to five of the herbicide-resistant plants were flagged per row, depending on the plant types observed. We selected 48 HR accessions to be characterized in terms of morphology (e.g. plant height, flag leaf length and width, panicle length, hull color, and awn length), phenology (days to heading), seed shattering, and GC (Tables I–V; Supplemental Table S2). Three reference cultivars (Clearfield hybrid rice cv CL XL745 and CL XL729 and Clearfield inbred rice cv CL151) were also characterized. Heading date was recorded when four tillers per plant had exerted panicles. Plant height was measured from the base of the culm to the tip of the flag leaf of the main culm. Leaf and panicle measurements were recorded from five tillers per plant. Awn length was measured for five seeds of the same panicle. Hull color was evaluated at harvest. At grain filling, the marked plants were enclosed in Delnet bags to catch any seeds that may shatter before harvest. A total of 735 plants were harvested 45 d from heading, over a 10-week period, from September 19 to November 23. At harvest, all tillers (still enclosed in Delnet bags) were cut manually at about 10 cm from the ground. A set of weights was attached to each bundle of tillers near the panicle end to achieve a total weight of 2.5 kg (plant material + added weight) per sample, which was intended to minimize the potential differences in air resistance (and, thus, maximum velocity) that might result for bundles of different sizes or shapes. Bundles were dropped panicle end down, from a height of 1 m, into a hard metal receptacle. Thus, the same force (approximately 50 kg force) and striking velocity {approximately 4.43 m s−1 [=9.8 × (√(2/9.8)]} were use to induce artificial shattering in all the samples. The shattered seeds were weighed. The panicles were threshed, the chaff was removed using a blower, and the remaining seed was weighed. The degree of shattering was calculated using the weight of shattered seeds versus the total weight of seeds per plant.
Seeds were afterripened at room temperature and germinated at 75 and 270 d from harvest; thus, samples were germinated in batches according to their harvest dates. Thirty seeds per characterized HR plant were placed in a petri dish (9 cm diameter) lined with filter paper and then moistened with 5 mL of deionized water. The dishes were placed in trays wrapped in plastic to prevent desiccation and dark incubated in a growth chamber at 30°C. Germinated seeds were counted, and removed, every 4 d up to 12 d of incubation. After each germination evaluation, the petri plates in each tray and the placement of trays in the growth chamber were rerandomized. Seeds were considered germinated when the radicle protruded from the caryopsis. At the end of the incubation period, nongerminated, intact seeds were counted and considered dormant based on previous tests (Tseng et al., 2013).
PROC CLUSTER analysis was conducted using SAS JMP Pro version 11 to determine accession groupings based on various response variables.
GBS
The 48 samples of HR weedy rice that were phenotyped were selected for genotyping (Supplemental Table S2). Also selected were 11 U.S. cultivars of the tropical japonica type (one of which is the inbred Clearfield reference line cv CL151), two Asian tropical japonica cultivars, 10 Asian indica cultivars, 10 Asian aus cultivars, four samples of wild species of Oryza, and 17 historical samples of U.S. weedy rice from Reagon et al. (2010), sampled prior to the widespread use of HR cultivars in the southern United States (Supplemental Table S3). These latter weeds were partitioned into 11 BHA weedy rice samples and six SH weedy rice samples. Approximately 100 mg of young and healthy leaf tissue was collected for each plant. Tissue was ground with a Retsch Mixer Mill MM400 with 3.2-mm stainless steel beads provided by BioSped Products. Total DNA was extracted with the Qiagen DNeasy Miniprep Kit following the manufacturer’s protocol. DNA was quantified with a Qubit2.0 Fluorometer. One microgram of high-quality DNA at 100 ng μL−1 for each sample was sent to the Cornell University Institute for Genomic Diversity (http://www.igd.cornell.edu/) for GBS (Elshire et al., 2011). Samples were digested with the restriction enzyme ApeKI, prepared following the protocol described by Elshire et al. (2011), and sequenced on an Illumina HiSeq. Raw reads are available under National Center for Biotechnology Information Short Read Archive experiment accession number SRX576894.
Reads were aligned to the MSU6 Nipponbare rice genome using the BWA software package (Li and Durbin, 2009). Initial data processing was performed at the Institute for Genomic Diversity at Cornell University with the Tassel pipeline (http://sourceforge.net/projects/tassel/), using the following parameters: a minimum minor allele frequency of 0.01 or greater (1%) was required for an SNP to be retained; a minimum locus coverage (proportion of taxa with a genotype) of 0.1 (10%) was required for retention; any SNP with more than two alleles was excluded.
Further filtering was done in our laboratory to remove SNPs that had more than 10% missing data and individuals with more than 95% missing data. We also removed any SNP involved in a long (more than 5 bp) mononucleotide repeat or immediately adjacent to one. Across the entire data set, 62,484 high-quality SNPs were obtained (Supplemental Data Set S1).
Population Structure Analyses
The population structure for various partitions of our GBS data set was determined using STRUCTURE 2.3.3 and an admixture model (Pritchard et al., 2000). A total of 10,000 loci were randomly chosen for each analysis to accommodate capacity limitations in STRUCTURE. Original analyses were run from K = 1 to K = 22, with four runs per K value, 100,000 burn-in period, and 500,000 replications on the Bioportal at the University of Oslo (http://www.bioportal.uio.no). Subsequent analyses containing only HR weeds and their immediate putative ancestral populations were run from K = 1 to K = 7, three runs per K value, and otherwise using the same parameters as above on the Massachusetts Green High Performance Computing Center cluster (http://www.mghpcc.org/). Due to the fact that cultivated and weedy rice are primarily inbreeding and mixed mating systems cannot be modeled in a STRUCTURE run, all runs were set with ploidy equal to 1. Heterozygous calls were coded as missing data. The best-fit K value was determined using Structure Harvester (Earl, 2012) and assessing ΔK and maximum likelihood scores (Supplemental Tables S4 and S5). A graphical representation of the population structure was generated using DISTRUCT (Rosenberg, 2004).
Principal component analysis was carried out with the smartpca program from EIGENSOFT 4.2 (Patterson et al., 2006; Price et al., 2006). The number of output eigenvectors was set to two, and the number of outlier iterations was set to five. Eigenvector graphing was carried out with RStudio (www.rstudio.com) for principal components 1 and 2. Wild rice samples were excluded from the principal component analysis.
Parental Contributions to HR Weedy Rice
We assessed for parental bias in the genomic contributions to HR weedy rice using FST and linkage disequilibrium methods. To decrease the probability of miscalled heterozygous sites leading to the incorrect assignment of ancestry to a given locus, GBS data were examined from sets of individuals in each of the three possible HR weed parental populations: tropical japonica, the historical BHA weed group, and the historical SH weed group. SNPs were dropped whenever one or more of the parental individuals were heterozygous at the site. Sites that contained heterozygous calls for HR weeds but not for any parental individuals were retained. Additionally, sites with calls of N (missing data) in the parental individuals did not lead to the exclusion of the SNP. A total of 17,562 SNPs were retained for parental contribution analyses.
We estimated SNP-specific FST between the historical and new HR weed populations and searched for outliers using the program LOSITAN. A total of 50,000 simulations were run using default parameters. Both the neutral mean FST and force mean FST options were used. Loci with high FST values between historical accessions and new HR weeds were considered highly differentiated and likely contributed by the tropical japonica parent to HR weeds. SNPs and FST values obtained are listed in Supplemental Table S6.
We further used the program LAMP-LD (Baran et al., 2012) to infer the ancestry of regions along each hybrid weed individual’s genome. The LAMP-LD algorithm uses a fast approximation of the model of Li and Stephens (2003). The genome is divided into windows that do not contain any transitions in ancestry. This corrects the tendency of the standard Li and Stephens (2003) model to predict artificially frequent transitions in local ancestry (Baran et al., 2012). Three ancestral populations were considered: tropical japonica, historical SH weeds, and historical BHA weeds. Inferences about the ancestry of each SNP were made in the HR weeds; in some cases, sites were inferred to have two ancestors (to be heterozygous), and we retained this inference. To assess the tendency for broad regions of the genome to be rich or poor in content from one of the three parental populations, we examined all HR weed SNPs and flagged those where at least 80% of the HR weed individuals carried at least one allele of any given parental origin. We then examined how many contiguous SNPs had similar high parental content. Low parental contribution was defined as SNPs and regions where less than 20% of the HR individuals carried alleles of a given parental origin. These definitions can and do overlap; for example, a region can have both high BHA contribution and low SH contribution. Chromosomal regions with contiguous runs of alleles stemming from one parental contributor are listed in Supplemental Table S7. The distribution of parental contribution in the population of HR weeds is shown in Figure 7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rice-producing counties in Arkansas, along the Mississippi River Delta, from where surviving or escaped weedy rice accessions were collected at the end of the 2010 cropping season.
Supplemental Table S1. Weedy rice accessions collected in 2010 from rice fields in Arkansas with a history of Clearfield rice.
Supplemental Table S2. Morphophysiological characteristics of herbicide-resistant weedy rice plants used in the GBS experiment.
Supplemental Table S3. Accessions characterized using GBS and used in the population structure analyses.
Supplemental Table S4. Structure Harvester (Earl, 2012) results for the STRUCTURE run including weedy, cultivated, and wild rice.
Supplemental Table S5. Structure Harvester (Earl, 2012) results for the STRUCTURE run including weedy and cultivated rice.
Supplemental Table S6. SNP-specific FST and heterozygosity and significance based on LOSITAN.
Supplemental Table S7. Chromosomal coordinates (MSU6 genome) of contiguous runs of high and low parental contribution in HR weeds.
Supplemental Data Set S1. SNPs obtained for rice samples using GBS.
Supplementary Material
Acknowledgments
We thank Ed Allan Alcober, Mariccor S.A.B. Batoy, George Botha, Leopoldo Estorninos, Jr., Reiofeli Salas, and Shilpa Singh for help in the characterization of plants.
Glossary
- HR
herbicide-resistant
- SH
straw-colored hulls
- BHA
black-colored hulls and awns
- DAP
days after planting
- GC
germination capacity
- GBS
genotyping by sequencing
- SNP
single-nucleotide polymorphisms
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1032023).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. (2008) LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi M, Azlan S, Yim KM, George TV, Chew SE. (2012) Control of weedy rice in direct-seeded rice using the Clearfield Production System in Malaysia. Pak J Weed Sci Res 18: 49–53 [Google Scholar]
- Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, Eng C, Rodriguez-Cintron W, Chapela R, Ford JG, Avila PC, et al. (2012) Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28: 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachell HM, Adair CR, Jodon NE, Davis LL, Jones JW. (1938) Extent of natural crossing in rice. J Am Soc Agron 30: 743–753 [Google Scholar]
- Beaumont MA, Nichols RA. (1996) Evaluating loci for use in the genetic analysis of population structure. Proc Biol Sci 263: 1619–1626 [Google Scholar]
- Burgos NR, Norman RJ, Gealy DR, Black H. (2006) Competitive N uptake between rice and weedy rice. Field Crops Res 99: 96–105 [Google Scholar]
- Burgos NR, Norsworthy JK, Scott RC, Smith KL. (2008) Red rice status after five years of Clearfield rice technology in Arkansas. Weed Technol 22: 200–208 [Google Scholar]
- Burgos NR, Scott RC, Guice JB. (2007) Detectable gene flow in commercial rice fields and impact of Clearfield technology on red rice infestation. In Proceedings of the 47th Annual Meeting. Weed Sci Soc Am. 47: 300 [Google Scholar]
- Burgos NR, Shivrain VK, Scott RC, Mauromoustakos A, Kuk YI, Sales MA, Bullington J. (2011) Differential tolerance of weedy red rice (Oryza sativa L.) from Arkansas, USA to glyphosate. Crop Prot 30: 986–994 [Google Scholar]
- Busconi M, Rossi D, Lorenzoni C, Baldi G, Fogher C. (2012) Spread of herbicide-resistant weedy rice (red rice, Oryza sativa L.) after 5 years of Clearfield rice cultivation in Italy. Plant Biol 14: 751–759 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, et al. (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet 3: 1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BS, Singh K, Ladha JK, Kumar V, Saharawat YS, Gathala M. (2013) Weedy rice: an emerging threat for direct-seeded rice production systems in India. J Rice Res 1: 106 [Google Scholar]
- Chen LJ, Lee DS, Song ZP, Suh HS, Lu BR. (2004) Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Annals Bot 93: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counce CE, Keisling TC, Mitchell AJ. (2000) A uniform, objective and adaptive system for expressing rice development. Crop Sci 40: 436–443 [Google Scholar]
- Craig SM, Reagon M, Resnick LE, Caicedo AL. (2014) Allele distributions at hybrid incompatibility loci facilitate the potential for gene flow between cultivated and weedy rice in the US. PLoS ONE 9: e86647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croughan TP. June 30, 1998. Herbicide resistant rice. US Patent Application No. 5,773,704
- Croughan TP. (2003) Clearfield rice: It’s not a GMO. Louisiana Agric 46: 24–26 [Google Scholar]
- Delouche JC, Burgos NR, Gealy DR, Zorilla-San Martin G, Labrada R, Larinde N (2007) Weedy Rices: Origin, Biology, Ecology and Control. Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- Diarra A, Smith RJ, Jr, Talbert RE. (1985) Interference of red rice (Oryza sativa) with rice (O. sativa). Weed Sci 33: 644–649 [Google Scholar]
- Do Lago A (1982) Characterization of red rice (Oryza sativa L.) phenotypes in Mississippi. PhD thesis. Mississippi State University, Starkville, MI [Google Scholar]
- Earl DA. (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4: 359–361 [Google Scholar]
- Eleftherohorinos I, Dhima KV, Vasilakoglou IB. (2002) Interference of red rice in rice grown in Greece. Weed Sci 50: 167–172 [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gealy DR (2005) Gene movement between rice (Oryza sativa) and weedy rice (Oryza sativa): a U.S. temperate rice perspective. In J Gressel, ed, Crop Ferality and Volunteerism. CRC Press, Boca Raton, FL, pp 323–354 [Google Scholar]
- Gealy DR, Agrama H, Jia MH. (2012) Genetic analysis of atypical U.S. red rice phenotypes: indications of prior gene flow in rice fields? Weed Sci 60: 451–461 [Google Scholar]
- Gealy DR, Agrama HA, Eizenga GC. (2009) Exploring genetic and spatial structure of U.S. weedy red rice (Oryza sativa) in relation to rice relatives worldwide. Weed Sci 57: 627–643 [Google Scholar]
- Gealy DR, Yan W, Rutger JN. (2006) Red rice (Oryza sativa) plant types affect growth, coloration, and flowering characteristics of first and second generation crosses with rice. Weed Technol 20: 839–852 [Google Scholar]
- Goss WL, Brown E. (1939) Buried red rice seed. J Am Soc Agron 31: 633–637 [Google Scholar]
- Gressel J, Valverde BE. (2009) A strategy to provide long-term control of weedy rice while mitigating herbicide resistance transgene flow, and its potential use for other crops with related weeds. Pest Manag Sci 65: 723–731 [DOI] [PubMed] [Google Scholar]
- Grimm NB, Faeth SH, Golubiewski NE, Redman CL, Wu J, Bai X, Briggs JM. (2008) Global change and the ecology of cities. Science 319: 756–760 [DOI] [PubMed] [Google Scholar]
- Kummu M, Ward PJ, de Moel H, Varis O. (2010) Is physical water scarcity a new phenomenon? Global assessment of water shortages over the last two millennia. Environ Res Lett 5: 034006 [Google Scholar]
- Kwon SL, Smith RJ, Jr, Talbert RE. (1991) Interference of red rice (Oryza sativa) densities in rice (O. sativa). Weed Sci 39: 197–174 [Google Scholar]
- Li H, Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. (2003) Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Schaal BA. (2007) Origins and population genetics of weedy red rice in the USA. Mol Ecol 16: 4523–4535 [DOI] [PubMed] [Google Scholar]
- Messeguer J, Marfa V, Catala MM, Guiderdoni E, Mele E. (2004) A field study of pollen-mediated gene flow from Mediterranean GM rice to conventional rice and the red rice weed. Mol Breed 13: 103–112 [Google Scholar]
- Noldin JA. (1995) Characterization, seed longevity, and herbicide sensitivity of red rice (O. sativa L.) ecotypes and red rice control in soybean [(Glycine max (L.) Merr.]. PhD thesis. Texas A&M University, College Station, TX
- Noldin JA, Yokoyama S, Antunes P, Luzzardi R. (2002) Outcrossing potential of glufosinate-resistant rice to red rice. Planta Daninha 20: 243–251 [Google Scholar]
- Ottis BV, Smith KL, Scott RC, Talbert RE. (2005) Rice yield and quality as affected by cultivar and red rice (Oryza sativa) density. Weed Sci 53: 499–504 [Google Scholar]
- Ouyang Y, Zhang Q. (2013) Understanding reproductive isolation based on the rice model. Annu Rev Plant Biol 64: 111–135 [DOI] [PubMed] [Google Scholar]
- Pandey S, Velasco L (2002) Economics of direct-seeding in Asia: patterns of adoption and research priorities. In S Pandey, M Mortimer, L Wade, TP Tuong, K Lopez, B Hardy, eds, Direct Seeding: Research Strategies and Opportunities. IRRI Press, Los Banos, Philippines, pp 3–14 [Google Scholar]
- Pantone DJ, Baker JB. (1991) Weed-crop competition models and response-surface analysis of red rice competition in cultivated rice: a review. Crop Sci 31: 1105–1110 [Google Scholar]
- Patterson N, Price AL, Reich D. (2006) Population structure and eigenanalysis. PLoS Genet 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AN, Johnson DE, Sivaprasad B, Ladha JK, Mortimer AM. (2007) Weed management in direct-seeded rice. Adv Agron 93: 153–255 [Google Scholar]
- Reagon M, Thurber CS, Gross BL, Olsen KM, Jia Y, Caicedo AL. (2010) Genomic patterns of nucleotide diversity in divergent populations of U.S. weedy rice. BMC Evol Biol 10: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagon M, Thurber CS, Olsen KM, Jia Y, Caicedo AL. (2011) The long and the short of it: SD1 polymorphism and the evolution of growth trait divergence in U.S. weedy rice. Molec Ecol 20: 3743–3756 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138 [Google Scholar]
- Sales MA, Shivrain VK, Burgos NR, Kuk YI. (2008) Amino acid substitutions in the acetolactate synthase gene of red rice (Oryza sativa) confer resistance to imazethapyr. Weed Sci 56: 485–489 [Google Scholar]
- Shimono H, Hasegawa T, Moriyama M, Fujimura S, Nagata T. (2005) Modeling spikelet sterility induced by low temperature in rice. Agron J 97: 1524–1536 [Google Scholar]
- Shivrain V, Burgos N, Anders M, Rajguru S, Moore J, Sales M. (2007) Gene flow between ClearfieldTM rice and red rice. Crop Prot 26: 349–356 [Google Scholar]
- Shivrain VK, Burgos NR, Agrama HA, Lawton-Rauh A, Lu BR, Sales MA, Boyett V, Gealy DR, Moldenhauer KAK. (2010a) Genetic diversity of weedy rice (Oryza sativa) in Arkansas, USA. Weed Res 50: 289–302 [Google Scholar]
- Shivrain VK, Burgos NR, Gealy DR, Moldenhauer KAK, Baquireza CJ. (2008) Maximum outcrossing rate and genetic compatibility between red rice (Oryza sativa) biotypes and Clearfield rice. Weed Sci 56: 807–813 [Google Scholar]
- Shivrain VK, Burgos NR, Gealy DR, Sales MA, Smith KL. (2009a) Gene flow from weedy red rice (Oryza sativa L.) to cultivated rice and fitness of hybrids. Pest Manag Sci 65: 1124–1129 [DOI] [PubMed] [Google Scholar]
- Shivrain VK, Burgos NR, Gealy DR, Smith KL, Scott RC, Mauromoustakos A, Black H. (2009b) Red rice (Oryza sativa L.) emergence characteristics and influence on rice (O. sativa L.) yield at different planting dates. Weed Sci 57: 94–102 [Google Scholar]
- Shivrain VK, Burgos NR, Moldenhauer KAK, McNew RW, Baldwin TL. (2006) Characterization of spontaneous crosses between Clearfield rice (Oryza sativa) and red rice (O. sativa). Weed Technol 20: 576–584 [Google Scholar]
- Shivrain VK, Burgos NR, Sales MA, Mauromoustakos A, Gealy DR, Smith KL, Black HL, Jia M. (2009c) Factors affecting the outcrossing rate between Clearfield rice and red rice (Oryza sativa). Weed Sci 57: 394–403 [Google Scholar]
- Shivrain VK, Burgos NR, Scott RC, Gbur EE, Jr, Estorninos LE, Jr, McClelland MR. (2010b) Phenotypic diversity of weedy red rice (Oryza sativa L.) in Arkansas, USA in relation to weed management. Crop Prot 29: 721–730 [Google Scholar]
- Song ZP, Lu BR, Zhu YG, Chen JK. (2003) Gene flow from cultivated rice to the wild species Oryza rufipogon under experimental field conditions. New Phytol 157: 67–665 [DOI] [PubMed] [Google Scholar]
- Sudianto E, Song BK, Neik TX, Saldain NE, Scott RC, Burgos NR. (2013) Clearfield rice: its development, success, and key challenges on a global perspective. Crop Prot 49: 40–51 [Google Scholar]
- Tan S, Evans RR, Dahmer ML, Singh BK, Shaner DL. (2005) Imidazolinone-tolerant crops: history, current status and future. Pest Manag Sci 61: 246–257 [DOI] [PubMed] [Google Scholar]
- Thurber C, Reagon M, Olsen KM, Jia Y, Caicedo AL. (2014) The evolution of flowering strategies in US weedy rice. Am J Bot 101: 1737–1747 [DOI] [PubMed] [Google Scholar]
- Thurber CS, Reagon M, Gross BL, Olsen KM, Jia K, Caicedo AL. (2010) Molecular evolution of shattering loci in U.S. weedy rice. Molec Ecol 19: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TM, Burgos NR, Shivrain VK, Alcober EA, Mauromoustakos A. (2013) Inter- and intrapopulation variation in dormancy of Oryza sativa (weedy red rice) and allelic variation in dormancy-linked loci. Weed Res 53: 440–451 [Google Scholar]
- Turral H, Burke J, Faures JM (2011) Climate Change, Water and Food Security. Food and Agriculture Organization of the United Nations, Rome
- Vidotto F, Ferrero A. (2000) Germination behaviour of red rice (Oryza sativa L.) seeds in field and laboratory conditions. Agronomie 20: 375–382 [Google Scholar]
- Waples RS, Gaggiotti O. (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15: 1419–1439 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Vaughan D, Tomooka N. (2000) Weedy rice complexes: case studies from Malaysia, Vietnam and Surinam. In B Baki, D Chin, M Mortimer, eds, Wild and Weedy Rice in Rice Ecosystems in Asia: A Review, Limited Proc #2. IRRI, Los Baños, Philippines, pp 25–34
- Zhang W, Linscombe SD, Webster E, Tan S, Oard J. (2006) Risk assessment of the transfer of imazethapyr herbicide tolerance from Clearfield rice to red rice (Oryza sativa). Euphytica 152: 75–86 [Google Scholar]
- Zong Y, Chen Z, Innes JB, Chen C, Wang Z, Wang H. (2007) Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449: 459–462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.