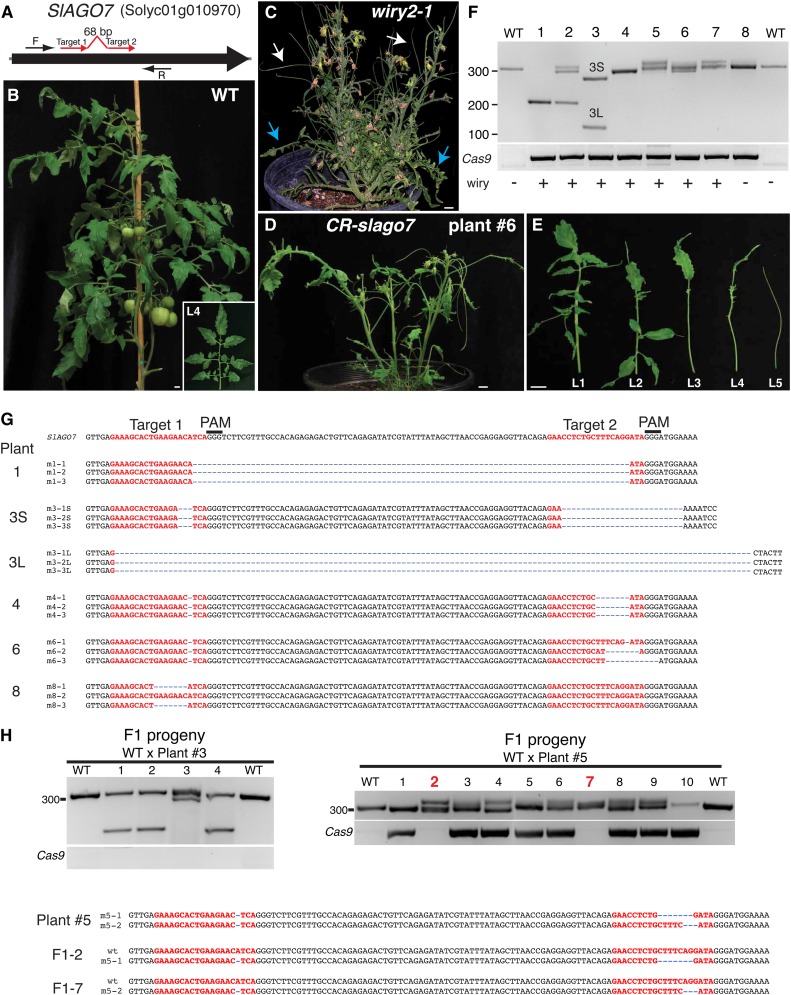
Figure 1.
CRISPR/Cas9-mediated gene editing in stable transgenic tomato plants. A, Schematic illustrating two sgRNAs (red arrows) targeting the SlAGO7 coding sequence. Cas9/sgRNA1/sgRNA2 were expressed from the same plasmid (Belhaj et al., 2013). Black arrows indicate PCR primers used to evaluate mutation type and efficiency. B, A wild-type (WT) tomato plant at 9 weeks of age, and the fourth-produced leaf from the primary shoot (inset). C, The strong wiry2-1 allele of slago7. First formed leaves have leaflets lacking petioles (blue arrows), and later formed leaves are radialized (white arrows). D and E, A representative CRISPR/Cas9-slago7 (CR-slago7) plant (D) and its first five leaves showing the distinctive loss-of-function recessive wiry syndrome (E). Bars = 1 cm. F and G, PCR genotyping of eight representative CR-slago7 plants showing the type of DNA lesions generated, including homozygosity for the expected 90-bp deletion (plant 1). PCR for the presence of Cas9 is also shown along with wiry phenotypic evaluations. A range of indels was found in the other T0 plants. The PCR products from a subset of plants were cloned and sequenced to validate the deletion in plant 1 and to characterize additional DNA lesions (G). Note that the two deletions in plant 3 are smaller (3S) and larger (3L) than expected. H, Germline transmission and heritability of the small and large deletions from plant 3 (left) and two small deletions from plant 5 (right) in the absence of the inductive Cas9/sgRNA1/sgRNA2 transgene. All four progeny from plant 3 lacked the CRISPR/Cas9 transgene, and all four were heterozygous for deletions, three of which were heterozygous for the large deletion and one heterozygous for the smaller deletion. Of the 10 progeny from plant 5, two plants lacked the CRISPR/Cas9 transgene, and sequencing showed that both were heterozygous for the wild-type allele and one of the two deletions (bottom).