FIGURE 1.
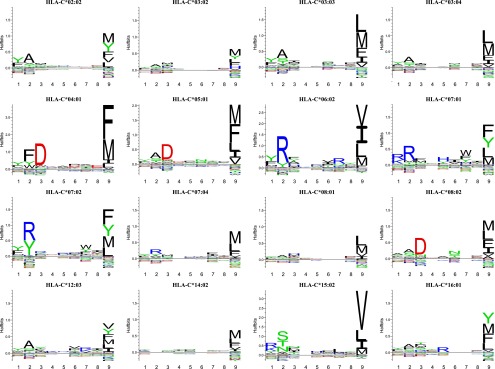
HLA-C allotypes exhibit classical MHC-I peptide-binding motifs. The peptide-binding motifs of 16 HLA-C allotypes were determined by SPA-driven PSCPL analysis using a nonameric peptide library. Binding motifs are represented as logos generated from peptide-binding matrices using the Seq2Logo server (24). The height of each letter indicates the information content, that is, larger letters indicate residues having more impact on binding in the given position, be it positive or negative. Amino acids plotted on the positive y-axis contribute positively to peptide binding, whereas amino acids plotted on the negative y-axis contribute negatively to peptide binding. The total height of a given position indicates the information content, and thus higher stacks represent amino acid position important for peptide binding. Each logo is an average of two independent experiments.
