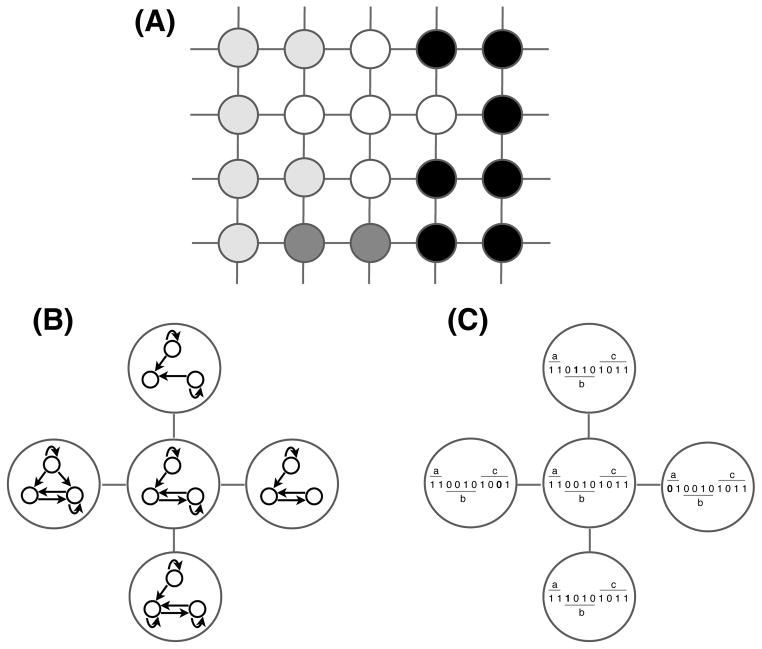
Figure 1.
(A) The space of all possible genotypes can be represented as a network, where vertices correspond to genotypes and edges connect genotypes that can be interconverted through point mutations. A genotype network is a connected subgraph in which each genotype yields the same phenotype (denoted by vertex color). Thus, all point mutations within a genotype network leave the phenotype unchanged. (B) When perturbations correspond to the addition or deletion of regulatory interactions, two RBCs are connected in a genotype network if they share the same gene expression pattern (phenotype), but differ in a single regulatory interaction. (C) When perturbations correspond to changes in the signal-integration logic, two RBCs are connected in a genotype network if they share the same phenotype, but differ in a single bit of their rule vector. This perturbation is analogous to a change in the affinity or position of a transcription factor binding site in the cis-regulatory region that leaves the gene expression pattern unchanged. The genotype networks shown in (B,C) are part of the larger (hypothetical) genotype space shown in (A). The number of neighbors per vertex is intentionally reduced for visual clarity.