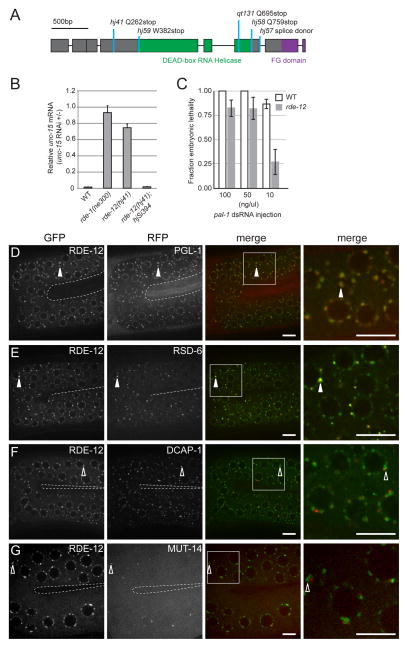
Figure 1. rde-12 is required for RNAi and is localized to P granules.
(A) Gene structure of rde-12. Exons are depicted in boxes. Mutant alleles and corresponding changes in the protein sequence are indicated. (B) Relative abundance of endogenous unc-15 mRNA in synchronized wild-type (WT), rde-1(ne300), rde-12(hj41) and rde-12(hj41); hjSi394[rde-12(+)] animals subjected to unc-15 RNAi compared to no RNAi control (L4440 vector). Mean + standard deviation from three independent samples assayed in triplicates are shown. (C) rde-12(qt131) animals were sensitive to RNAi against pal-1 when high doses of dsRNA were directly injected into the gonad. Number of injected animals n=8–17. Mean ± standard deviation. At 100 and 50ng/μl, wild type and rde-12 mutant animals were not significantly different. At 10ng/μl, p<0.01. (D–G) The germline (in the loop region connecting the proximal and distal gonad arms) of live animals were imaged by confocal microscopy. The inner border of gonad arms is depicted by dotted lines. Full-length RDE-12 was fused to GFP (GFP::RDE-12). (D) mRuby::PGL-1; (E) tagRFP::RSD-6; (F) mRuby::DCAP-1; (G) MUT-14::mCherry. Solid arrowheads indicate co-localization of GFP with RFP signals. Open arrowheads indicate the positions of GFP signals that do not overlap with RFP signals. Boxed areas were magnified 3x and shown in the far right panels. Scale bars, 6.5μm.