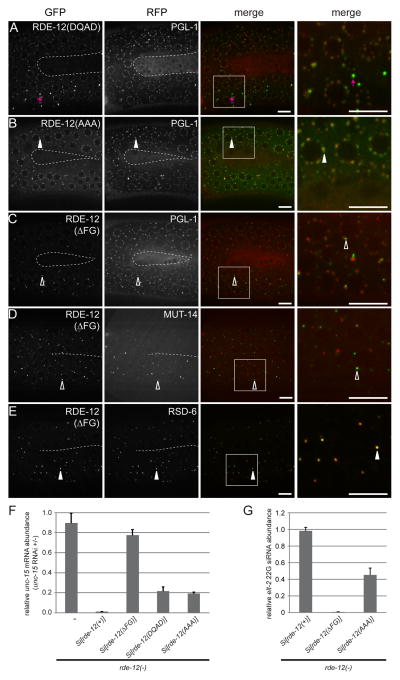
Figure 2. The FG domain is required for proper RDE-12 localization.
(A–E) The germline (in the loop region connecting the proximal and distal gonad arms) of live animals were imaged by confocal microscopy. The inner border of gonad arms is depicted by dotted lines. Boxed areas were magnified 3x and shown in the far right panels. (A–C) mRuby::PGL-1 and translational fusion of GFP with mutant forms of RDE-12. (A) DEAD motif mutated to DQAD, abnormal GFP aggregates are indicated by magenta arrows; (B) SAT motif mutated to AAA; (C) FG domain (amino acid residues 875-959) deleted. (D) GFP::RDE-12(ΔFG) and MUT-14::mCherry (Mutator foci marker). (E) GFP::RDE-12(ΔFG) and RSD-6. Solid arrowheads indicate co-localization of GFP with RFP signals. Open arrowheads indicate the positions of GFP signals that do not overlap with RFP signals. Boxed areas were magnified 3x and shown in the far right panels. Scale bars, 6.5μm. (F) Relative abundance of endogenous unc-15 mRNA in synchronized rde-12(hj41) mutant animals carrying the indicated transgeneswhen they were subjected to unc-15 RNAi compared to no RNAi control (L4440 vector). Mean + standard deviation from three independent samples assayed in triplicates is shown. (G) Relative abundance of elt-2 22G secondary siRNA in synchronized rde-12(hj41) mutant animals carrying the indicated transgenes when they were subjected to elt-2 RNAi. Mean + standard deviation from three independent samples assayed in triplicates is shown.