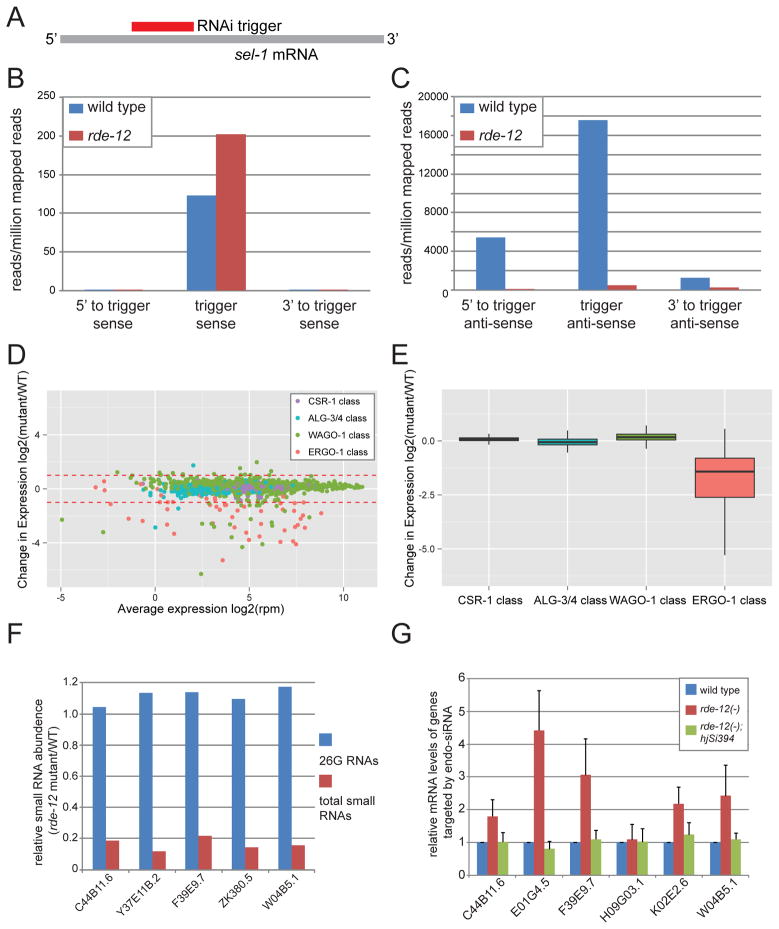
Figure 4. RDE-12 is required for secondary siRNA synthesis.
(A) Schematic representation of the sel-1 RNAi trigger and sel-1 mRNA. (B–C) The mean abundance of small RNAs from two independent populations of wild-type and rde-12(hj41) animals undergoing sel-1 RNAi is shown. (B) Sense sel-1 siRNAs. (C) Antisense sel-1 siRNAs. (D) MA plot showing abundance change of endo-siRNA in individual targets of ALG-3/4, CSR-1, ERGO-1 and WAGO-1. (E) Relative abundance of endo-siRNAs in wild-type (WT) versus rde-12 mutant animals. (F) The relative abundance of 26G RNAs and total endo-siRNAs in target genes of the somatic ERGO-1 pathway (WT=1.0). All listed target genes have >1 RPM 26G RNAs. (G) Relative abundance of endogenous mRNAs targeted by the ERGO-1 class 26G siRNAs in wild type, rde-12(hj41) and rde-12(hj41);hjSi394[rde-12(+)] animals as determined by real-time PCR (WT = 1.0).