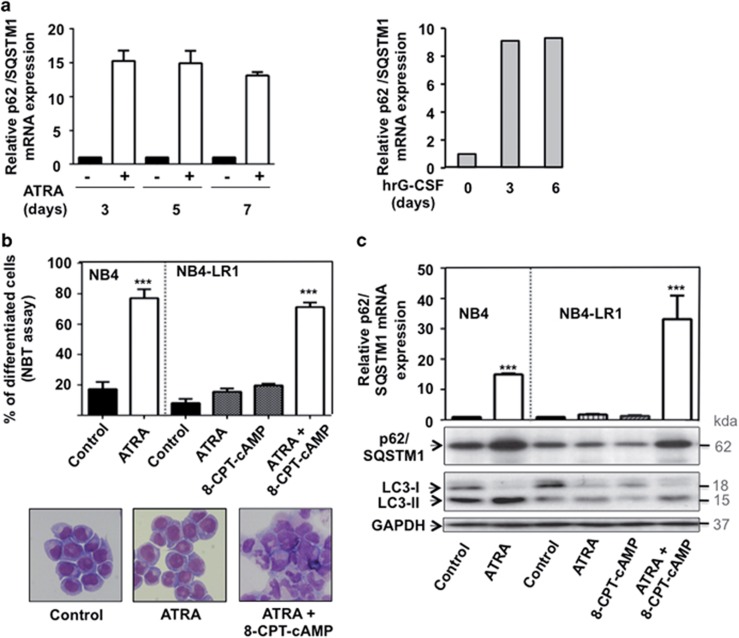
Figure 2.
ATRA-induced upregulation of p62/SQSTM1 mRNA occurs in NB4 maturation-sensitive cells but not in NB4-LR1 maturation-resistant cells. (a) NB4 cells (left panel) and primary CD34+progenitors cells (right panel) were treated with 1 μM ATRA and 10 ng/ml (hrG-CSF) for the indicated times, respectively. The p62/SQSTM1 mRNA levels were assessed, after RNA extraction, by RT-qPCR as described in Djavaheri-Mergny et al.56 (b and c) NB4-LR1 cells, which are resistant to maturation by ATRA, were treated with 1 μM ATRA and 100-μM 8-CPT-cAMP alone or in combination for 4 days. (b) Granulocytic differentiation was determined by FACS analysis of the expression CD11c or by morphologic changes of cells stained with May-Grunwald Giemsa. (c) Quantification of p62/SQSTM1 mRNA levels by RT-qPCR (upper panel) and immunoblot analysis of p62/SQSTM1 and LC3 proteins (lower panel). The relative expression of p62/SQSTM1 mRNA transcripts to the housekeeping genes, CPB (Figure a, left panel and Figure c) or HMBS (Figure a, right panel) was calculated as 2−ΔΔCT. For each cDNA, results were expressed as fold changes compared with untreated cells. ***P<0.001 versus untreated cells