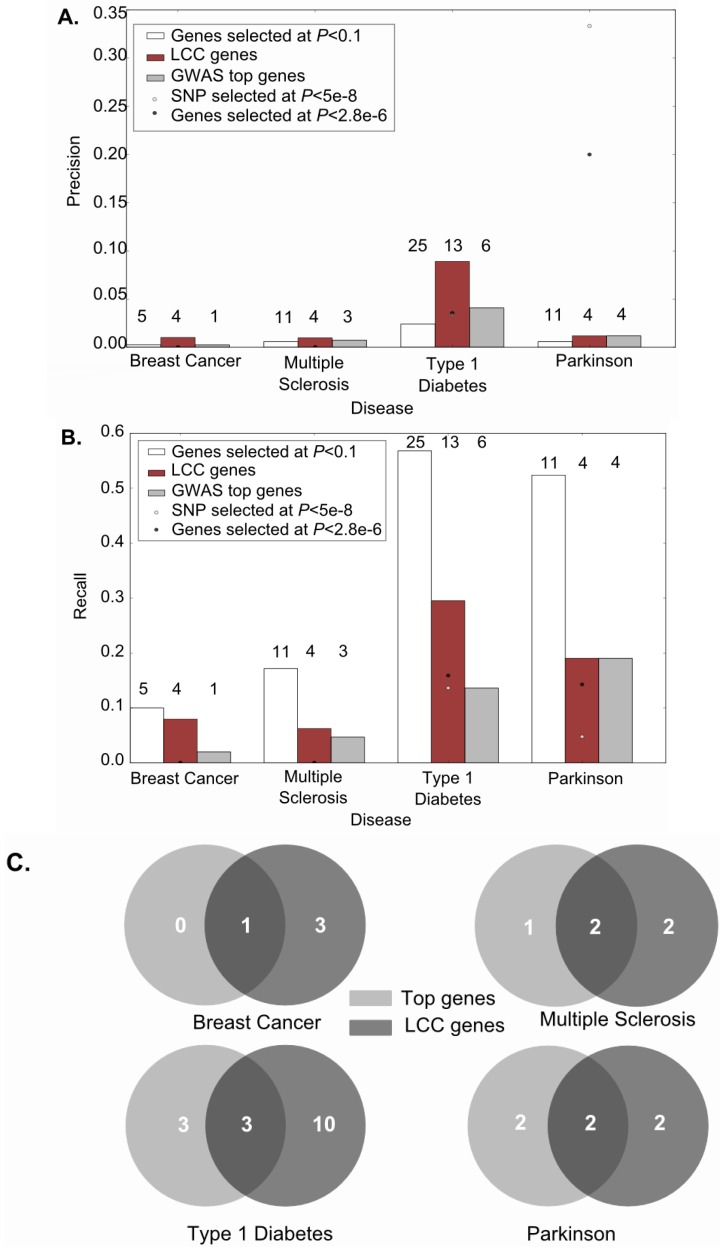
Figure 2.
Largest connected components contain true biological insight into diseases. (A) Precision, by disease, of five sets of genes against a list of known diseases candidates (n = 50, 64, 44 and 21 for breast cancer, multiple sclerosis, Parkinson’s and type 1 diabetes, respectively). The sets of genes evaluated for precision against the lists of known candidates were: the set of genes selected at a gene wise p-value cutoff of 0.1 (white bar) (n = 1934, 1894, 1035, 1907 in breast cancer, MS, Parkinson’s and type 1 diabetes, respectively), the set of genes included in the LCC obtained from the previous selection (red bar) (n = 395, 410, 146 and 337 in breast cancer, MS, Parkinson’s and type 1 diabetes, respectively), the same number of GWAS top genes than the ones included in the LCC (grey bar), the set of genes surviving Bonferroni correction over SNPs or genes (grey and black dots, respectively). Numbers above the bars are the number of known candidates included in each gene selection set; (B) Recall, by disease, of the same sets of genes against the lists of known candidates. Numbers above the bars are the number of known candidates included in each gene selection set; and (C) Venn diagrams showing, for each disease, the overlap between known candidate genes retrieved by LCC genes (dark grey circle) and by the same number of GWAS top genes (light grey circle).