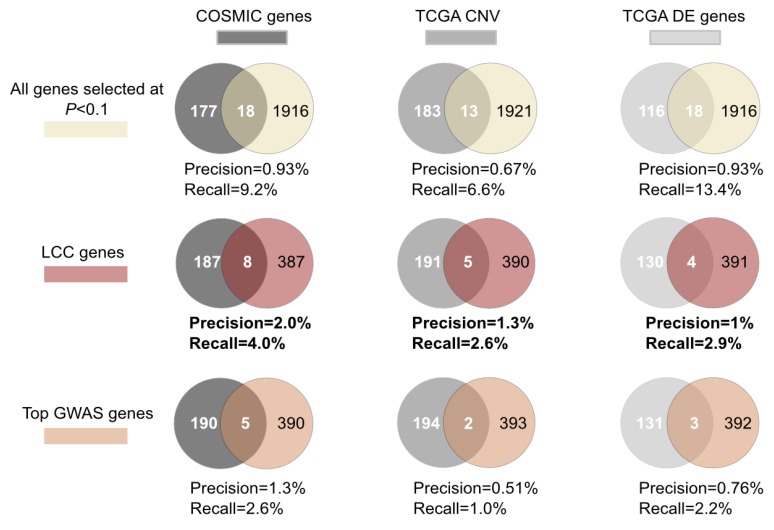
Figure 3.
LCC performs better than GWAS top genes in retrieving known breast cancer genes. Venn diagrams showing the overlap between genes reported to be differentially expressed (retrieved from the TCGA data portal, using the default parameters (−0.5 < Log2 < 0.5; frequency = 40%) or to harbor copy number abnormalities (retrieved from the TCGA data portal, using the default parameters −0.5 < Log2 < 0.5; frequency = 20%) or somatic mutation (genes with at least five cases reported in COSMIC database) in breast cancer, with each of the sets of genes selected from the breast cancer GWAS dataset by the previous gene selection approaches (all genes selected at a gene-wise p-value <0.1, LCC genes and top GWAS genes, represented in light yellow, red and orange circles, respectively).