Figure 3.
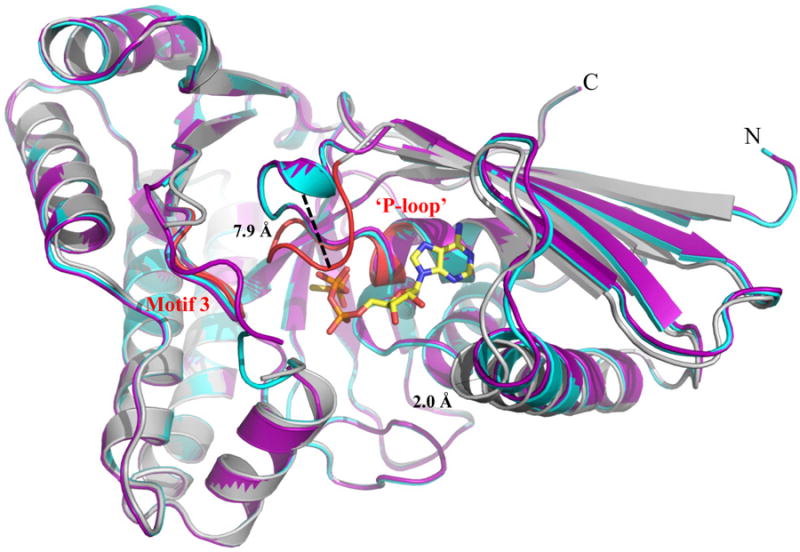
Structural alignment of WT MDD crystal structures. Cartoon ribbon diagram of the structural alignment of apo-MDD (purple), MDD bound to the substrate MVAPP (cyan) and MDD bound to the inhibitor FMVAPP and nucleotide analog ATPγS (gray). Only ATPγS is shown for clarity in ball and stick format (yellow). Conserved GHMP kinase family motifs 2 (‘P-loop’) and 3 are colored red within the ternary MDD co-crystal structure. A 7.9 Å shift within the ‘P-loop’, as measured from the Cα of Ala110 on both the apo-MDD and ternary MDD crystal structures, is highlighted for clarity. RMSD values for the structural alignment of all three structures can be found in Supplemental Table 1.
